Macrophages are present in all body tissues. They play a key role in the clearance of pathogens, participate in the immune and inflammatory responses, and partake in tissue repair and homeostasis. However, tissue infiltration of macrophages also exacerbates pathological processes, such as cancers [1-4]. Tumor-associated macrophages (TAMs) mostly originate from blood monocytes [5]. They are recruited to the tumor stroma at all stages of cancer progression [2,6] and can represent more than fifty percent of the tumor mass, thus representing by far the most abundant immune cell of the tumor stroma. Their number positively correlates with poor prognoses in most solid cancers as they are involved in several cancer-promoting events such as angiogenesis, lymphangiogenesis, immunosuppression, metastasis formation and resistance to therapies [7,8]. Therefore, the control of TAM infiltration is a current therapeutic strategy against cancers [9-11]. However, many questions regarding the mechanism of TAM tissue migration remain unresolved, which further hinders the development of novel therapeutic approaches.
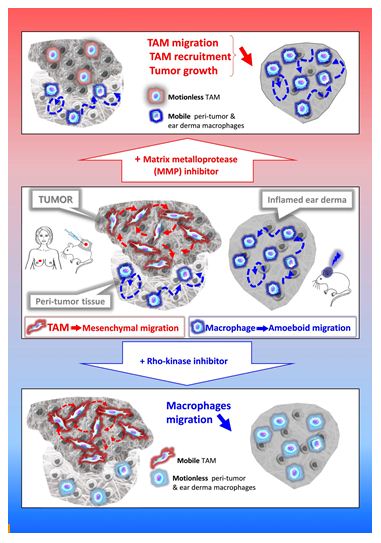
Cell migration in tissues occurs in three dimensions (3D) that profoundly differs from 2D migration processes [12,13]. Two main mechanistically distinct migration modes have been described in 3D environments: amoeboid and mesenchymal [14]. The amoeboid movement is characterized by rounded, ellipsoid, or moderately elongated cells that form blebs or generate small actin-rich filopodia [15-17]. These cells do not require adhesion to the extracellular matrix (ECM), but rather use a propulsive and pushing migration mode [16,18,19]. This non-directional motility involves acto-myosin contractions and depends on the Rho–ROCK pathway. Cells migrating through the mesenchymal mode adopt an elongated and protrusive morphology [15,17,18]. The movement is directional, involves cell adhesion to the substratum, and requires proteases to degrade the ECM in order to create paths through dense environments. In macrophages, in contrast to the amoeboid movement, mesenchymal migration is not inhibited, but rather stimulated, by treatment with ROCK inhibitors [20,21]. Unlike lymphocytes, neutrophils and monocytes [3,22,23], macrophages share the capacity with only few cell types including tumor cells or immature dendritic cells (DCs) [17,24,25], to use both amoeboid and mesenchymal migration modes in 3D environments. In vitro studies revealed that macrophages tailor their migration mode to the architecture of the surrounding ECM [20,22,26-35]. In vivo in mouse tumors and ex vivo in human breast cancer explants, macrophages use the two migration modes depending on the tissue they infiltrate. TAMs use the protease-dependent mesenchymal migration mode in mouse fibrosarcoma in vivo or human breast cancers ex vivo [20]. In contrast, in non-tumorous tissues such as the tumor periphery or in inflamed ear derma, macrophages use the amoeboid motility in vivo [20]. A chronic treatment with a broad-spectrum inhibitor of matrix metalloproteinases (MMPs) blocked the mesenchymal migration of macrophages, which correlates with a decrease in both TAM infiltration and tumor growth in vivo [20]. These findings strongly suggest that inhibition of TAM motility could be a way to impede their pro-tumor action and urge to identify specific effectors of the TAM mesenchymal migration as new targets in anti-cancer therapy.
Among effectors of TAM mesenchymal migration, MMPs need to be considered. MMP inhibitors have already been used to hamper tumor cell invasiveness by impeding tumor stroma remodeling and cancer cell escape from the primary tumor as well as decreasing angiogenesis [36,37]. Anti-tumor action of MMP inhibitors can now be explained by their action on TAM motility. Batimastat as well as its orally bioavailable derivative Marimastat were the first MMP inhibitors to enter clinical trials more than a decade ago [36]. However, clinical trials in patients with pancreatic, brain, lung or renal cancers were disappointing, essentially because these drugs were only tested in patients with advanced diseases despite the fact that studies in animal models had shown a most effective effect in treating early-stage diseases [38]. In addition, the primarily tested broad-spectrum MMP inhibitors were non-specific and did not differentiate between pro-tumor and anti-tumor MMPs depending on the type of cancer [38]. Thus, the recent knowledge on MMP biology and their differential involvement in tumor progression [39] together with the development of new generation MMP inhibitors [40-42] and the involvement of MMP activity on TAM motility stress the need to reassess the use of such inhibitors in early cancer treatment in combination with other anti-cancer molecules.
Another future strategy to identify new potential therapeutic targets consists in identifying new specific effectors of TAM mesenchymal migration. Therefore, exhaustive approaches to reach a comprehensive understanding of this process will be necessary as recently described in a transcriptomic-based analysis [27]. This strategy leads to the identification of a large number of potential targets and the future challenge will be to validate or invalidate all the potential hits as effective actors of macrophage migration both in vitro and in vivo through functional studies. For such large-scale screening approaches, new cellular tools are needed. Many studies use bone marrow-derived macrophages (BMDMs) from wild-type (WT) and knock-out (KO) mice or macrophage cell lines such as murine Raw 264.7 cells or human U937, HL-60 or THP1 cells for this purpose. All these cell models have several drawbacks such as the use of numerous animals, the limited number of cells and the impossibility to generate stable mutants in primary cultures or the fact that macrophage cell lines are usually distantly related to blood-derived macrophages or BMDMs particularly because they are cancer cells. Expansion of murine hematopoietic precursors that were transiently immortalized through a retroviral-delivery of an estrogen-inducible form of the transcription factor Hoxb8 has been described [43] and validated for the study of hematopoietic cell biology [43-52]. The possibility to use the CRISPR/Cas9 technology in this long-term hematopoietic progenitor cell lines has enabled the creation of new genetically modifiable cell models [45]. During the last few years, ectopic expression of Hoxb8 has been used in several studies mainly focused on DC biology [45,47,52-54], but also to generate surrogate macrophages [43,52,55]. This new cellular tool that combines the unlimited proliferative capacity of conditionally Hoxb8-immortalized hematopoietic progenitor cells with the CRISPR/Cas9 technology represents a powerful tool to genetically manipulate macrophages and explore their functionalities in a broad range of applications [55].
In conclusion, the tissue migration of TAMs emerges as a new therapeutic target to combat cancer diseases and the development of new cellular models to molecularly dissect the mesenchymal migration process should lead to the identification of new leads in anti-cancer therapy in the coming years.
References
- Allavena P, Chieppa M, Bianchi G, M Fabbri, G Laskarin, et al. (2010) Engagement of the mannose receptor by tumoral mucins activates an immune suppressive phenotype in human tumor-associated macrophages. Clinical & Developmental Immunology 2010: 547179.
- Condeelis J, Pollard JW (2006) Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124: 263-266. [crossref]
- Friedl P, Weigelin B (2008) Interstitial leukocyte migration and immune function. Nature Immunology 9: 960-969. [crossref]
- Yiangou Y, Facer P, Durrenberger P, Iain P Chessell, Alan Naylor, et al. (2006) COX-2, CB2 and P2X7-immunoreactivities are increased in activated microglial cells/macrophages of multiple sclerosis and amyotrophic lateral sclerosis spinal cord. BMC Neurology 6: 12. [crossref]
- Lahmar Q, Keirsse J, Laoui D, Kiavash Movahedi, Eva Van Overmeire, et al. (2016) Tissue-resident versus monocyte-derived macrophages in the tumor microenvironment. Biochim Biophys Acta 1865: 23-34. [crossref]
- Allavena P, Sica A, Solinas G, Chiara Porta, Alberto Mantovani (2008) The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Critical Reviews in Oncology/Hematology 66: 1-9. [crossref]
- Noy R, Pollard JW (2014) Tumor-associated macrophages: from mechanisms to therapy. Immunity 41: 49-61. [crossref]
- Ruffell B, Coussens LM (2015) Macrophages and therapeutic resistance in cancer. Cancer Cell 27: 462-472. [crossref]
- Mantovani A, Marchesi F, Malesci A, Luigi Laghi, Paola Allavena (2017) Tumour-associated macrophages as treatment targets in oncology. Nature Reviews Clinical Oncology 14: 399-416. [crossref]
- Morrison C (2016) Immuno-oncologists eye up macrophage targets. Nature reviews Drug discovery 15: 373-374. [crossref]
- Ostuni R, Kratochvill F, Murray PJ, Natoli G (2015) Macrophages and cancer: from mechanisms to therapeutic implications. Trends in Immunology 36: 229-239. [crossref]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM (2009) One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol 184: 481-490. [crossref]
- Hooper S, Marshall JF, Sahai E (2006) Tumor cell migration in three dimensions. Methods in Enzymology 406: 625-643.
- Van Goethem E, Le Cabec V, Poincloux R, Fabienne Gauffre, Isabelle Maridonneau-Parini (2010) Microenvironment structural constraints impact the 3D migration mode of macrophages and the formation of 3D podosomes. Eur J Clin Invest 40: 43.
- Friedl P, Wolf K (2003) Proteolytic and non-proteolytic migration of tumour cells and leucocytes. Biochem Soc Symp 277-85. [crossref]
- Lammermann T, Sixt M (2009) Mechanical modes of ‘amoeboid’ cell migration. Current Opinion in Cell Biology 21: 636-644. [crossref]
- Sahai E, Marshall CJ (2003) Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nature Cell Biology 5: 711-719. [crossref]
- Fackler OT, Grosse R (2008) Cell motility through plasma membrane blebbing. J Cell Biol 181: 879-884. [crossref]
- Paluch EK, Aspalter IM, Sixt M (2016) Focal Adhesion-Independent Cell Migration. Annu Rev Cell Dev Biol 32: 469-490. [crossref]
- Gui P, Ben-Neji M, Belozertseva E, Florence Dalenc, Camille Franchet, et al. (2018) The Protease-Dependent Mesenchymal Migration of Tumor-Associated Macrophages as a Target in Cancer Immunotherapy. Cancer Immunology Research 6: 1337-1351. [crossref]
- Gui P, Labrousse A, Van Goethem E (2014) Rho/ROCK pathway inhibition by the CDK inhibitor p27kip1 participates in the onset of macrophage 3D-mesenchymal migration. Journal of Cell Science 127: 4009-4023.
- Cougoule C, Van Goethem E, Le Cabec V, Fanny Lafouresse, Loïc Dupré, et al. (2012) Blood leukocytes and macrophages of various phenotypes have distinct abilities to form podosomes and to migrate in 3D environments. European Journal of Cell Biology 91: 938-949. [crossref]
- Lammermann T, Germain RN (2014) The multiple faces of leukocyte interstitial migration. Seminars in Immunopathology 36: 227-251. [crossref]
- Cougoule C, Lastrucci C, Guiet R, Rémi Mascarau, Etienne Meunier, et al. (2018) Podosomes, But Not the Maturation Status, Determine the Protease-Dependent 3D Migration in Human Dendritic Cells. Frontiers in immunology 9: 846. [crossref]
- Friedl P, Wolf K (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Reviews Cancer 3: 362-374. [crossref]
- Barros-Becker F, Lam PY, Fisher R, Huttenlocher A (2017) Live imaging reveals distinct modes of neutrophil and macrophage migration within interstitial tissues. Journal of CELL science 130: 3801-3808. [crossref]
- Cermak V, Gandalovicova A, Merta L, Jitka Fučíková, Radek Špíšek, et al. (2018) RNA-seq of macrophages of amoeboid or mesenchymal migratory phenotype due to specific structure of environment. Scientific Data 5: 180198. [crossref]
- Cougoule C, Le Cabec V, Poincloux R, Talal Al Saati, Jean-Louis Mège, et al. (2010) Three-dimensional migration of macrophages requires Hck for podosome organization and extracellular matrix proteolysis. Blood 115: 1444-1452. [crossref]
- Gui P, Labrousse A, Van Goethem E (2014) Rho/ROCK pathway inhibition by CDK inhibitor p27kip1 participates in the onset of macrophage 3D-mesenchymal migration. Journal of Cell Science.
- Guiet R, Verollet C, Lamsoul I, Céline Cougoule, Renaud Poincloux, et al. (2012) Macrophage mesenchymal migration requires podosome stabilization by filamin A. The Journal of Biological Chemistry 287: 13051-13062. [crossref]
- Jevnikar Z, Mirkovic B, Fonovic UP, Nace Zidar Urban Švajger, et al. (2012) Three-dimensional invasion of macrophages is mediated by cysteine cathepsins in protrusive podosomes. European Journal of Immunology 42: 3429-3441.
- Van Goethem E, Guiet R, Balor S, Guillaume M.Charrière, Renaud et al. (2011) Macrophage podosomes go 3D. European Journal of Cell Biology 90: 224-236.
- Van Goethem E, Poincloux R, Gauffre F, Isabelle Maridonneau-Parini, Véronique Le Cabec (2010) Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol 184: 1049-1061. [crossref]
- Lugo-Villarino G, Verollet C, Maridonneau-Parini I, Neyrolles O (2011) Macrophage Polarization: Convergence Point Targeted by Mycobacterium Tuberculosis and HIV. Frontiers in Immunology 2: 43. [crossref]
- Verollet C, Souriant S, Bonnaud E, Paul Jolicoeur, Brigitte Raynaud-Messina et al. (2015) HIV-1 reprograms the migration of macrophages. Blood 125: 1611-1622. [crossref]
- Hidalgo M, Eckhardt SG (2001) Development of matrix metalloproteinase inhibitors in cancer therapy. Journal of the National Cancer Institute 93: 178-193. [crossref]
- Amorosa V, Tebas P (2006) Bone disease and HIV infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 42: 108-114.
- Overall CM, Kleifeld O (2006) Tumour microenvironment – opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nature Reviews CANCER 6: 227-239. [crossref]
- Mason SD, Joyce JA (2011) Proteolytic networks in cancer. Trends in Cell Biology 21: 228-237. [crossref]
- Devel L, Czarny B, Beau F, Dimitris Georgiadis, Enrico Stura, et al. (2010) Third generation of matrix metalloprotease inhibitors: Gain in selectivity by targeting the depth of the S1′ cavity. Biochimie 92: 1501-1508. [crossref]
- Fingleton B (2008) MMPs as therapeutic targets–still a viable option? Seminars in Cell & Developmental Biology 19: 61-68. [crossref]
- Tauro M, McGuire J, Lynch CC (2014) New approaches to selectively target cancer-associated matrix metalloproteinase activity. Cancer Metastasis Rev 33: 1043-1057. [crossref]
- Wang GG, Calvo KR, Pasillas MP, David B Sykes, Hans Häcker, et al. (2006) Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods 3: 287-293. [crossref]
- Lee S, Kivimae S, Szoka FC (2017) Clodronate Improves Survival of Transplanted Hoxb8 Myeloid Progenitors with Constitutively Active GMCSFR in Immunocompetent Mice. Molecular Therapy Methods & Clinical Development 7: 60-73. [crossref]
- Hammerschmidt SI, Werth K, Rothe M, Melanie Galla, Marc Permanyer, et al. (2018) CRISPR/Cas9 Immunoengineering of Hoxb8-Immortalized Progenitor Cells for Revealing CCR7-Mediated Dendritic Cell Signaling and Migration Mechanisms in vivo. Frontiers in Immunology 9: 1949. [crossref]
- Di Ceglie I, van den Akker GG, Ascone G, Bas Ten Harkel, Hans Häcker, et al. (2017) Genetic modification of ER-Hoxb8 osteoclast precursors using CRISPR/Cas9 as a novel way to allow studies on osteoclast biology. Journal of Leukocyte Biology 101: 957-966. [crossref]
- Rosas M, Osorio F, Robinson MJ, Luke C Davies, Nicola Dierkes, et al. (2011) Hoxb8 conditionally immortalised macrophage lines model inflammatory monocytic cells with important similarity to dendritic cells. European Journal of Immunology 41: 356-365. [crossref]
- Chu JY, McCormick B, Mazelyte G, Melina Michael, Sonja Vermeren, et al. (2019) HoxB8 neutrophils replicate Fcgamma receptor and integrin-induced neutrophil signaling and functions. Journal of Leukocyte Biology 105: 93-100. [crossref]
- Witschi R, Johansson T, Morscher G, Louis Scheurer, Jacqueline Deschamps, et al. (2010) Hoxb8-Cre mice: A tool for brain-sparing conditional gene deletion. Genesis 48: 596-602. [crossref]
- Gurzeler U, Rabachini T, Dahinden CA, M Salmanidis, G Brumatti, et al. (2013) In vitro differentiation of near-unlimited numbers of functional mouse basophils using conditional Hoxb8. Allergy 68: 604-613. [crossref]
- Zach F, Mueller A, Gessner A (2015) Production and Functional Characterization of Murine Osteoclasts Differentiated from ER-Hoxb8-Immortalized Myeloid Progenitor Cells. PloS one 10: e0142211. [crossref]
- Cabron AS, El Azzouzi K, Boss M, Philipp Arnold, Jeanette Schwarz, et al. (2018) Structural and Functional Analyses of the Shedding Protease ADAM17 in HoxB8-Immortalized Macrophages and Dendritic-like Cells. J Immunol 201: 3106-3118. [crossref]
- Grajkowska LT, Ceribelli M, Lau CM, Margaret E Warren, Ioanna Tiniakou, et al. (2017) Isoform-Specific Expression and Feedback Regulation of E Protein TCF4 Control Dendritic Cell Lineage Specification. Immunity 46: 65-77. [crossref]
- Leithner A, Renkawitz J, De Vries I, Robert Hauschild, Hans Häcker, et al. (2018) Fast and efficient genetic engineering of hematopoietic precursor cells for the study of dendritic cell migration. European Journal of Immunology 48: 1074-1077. [crossref]
- Accarias S, Sanchez T, Labrousse A, Myriam Ben-Neji, Aurélien Boyance, et al. (2020) Genetic engineering of hoxb8 immortalized hematopoietic progenitors: a potent tool to study macrophage tissue migration. Journal of cell science.