DOI: 10.31038/CST.2019411
In addition to curing Wilm’s tumor and choriocarcinoma, I propose actinomycin to be more widely used to cure ALL cancer – i.e., curing lung, colon, breast, ovarian and other cancers – for the reasons described in this communication.
The discovery that actinomycin is a powerful anticancer agent against Wilm’s tumor and choriocarcinoma was made by Sidney Farber in the early 1950’s. This miraculous discovery is known to dramatically cure patients suffering from these embryonic tumors — and still remains the treatment of choice to this day!
Although it is not known whether actinomycin can be used to treat ALL cancers – the purpose of this letter is to call attention to the possibility that trace amounts of actinomycin given over an extended period of time could prove to be a powerful anticancer chemotherapeutic regimen. It is important that the medical community realize this, and to understand how actinomycin acts to defeat cancer.
Background Information
Actinomycin D is a naturally occurring cyclic-polypeptide containing antibiotic known to bind to DNA and inhibit RNA synthesis (see Figure 1) 1–4]. It does this by interfering with the elongation of growing RNA chains by the RNA polymerase enzyme 5]. Nucleolar 5S ribosomal RNA-synthesis is known to be particularly sensitive to the presence of actinomycin, and this accounts for its pharmacological activity as well as its extreme toxicity to mammalian cells [6, 7].
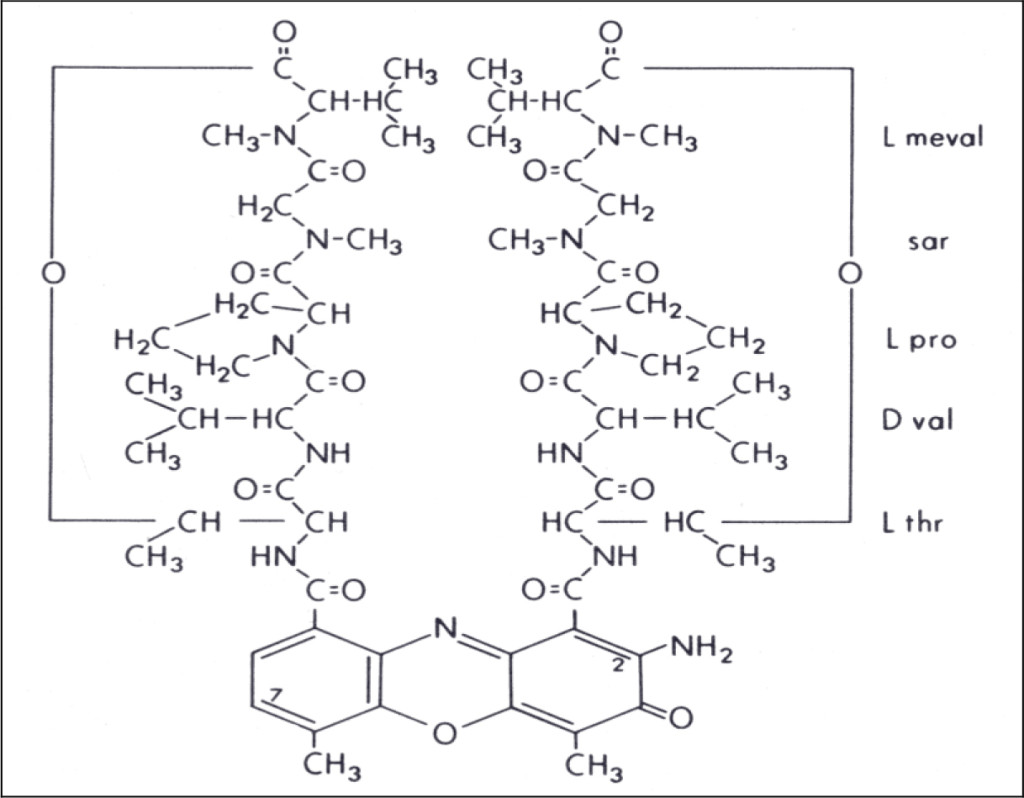
Figure 1. Chemical structure of actinomycin D. (L meval: L methyl valine; sar: sarcosine; L pro: L proline; D val: D valine; L thr: L threonine)
Stereochemistry of Actinomycin-DNA Binding
Several years ago, we determined the three-dimensional structure of an actinomycin-deoxyguanosine complex by x-ray crystallography [8–11]. The stereochemical information obtained from this study suggested a model to understand the general features of how actinomycin binds to DNA. According to this model, the phenoxazone ring system on actinomycin intercalates between adjacent guanine-cytosine base-pairs, while pentapeptide chains lie in the narrow groove of the B- helix to form hydrogen bonds with guanine residues on opposite chains. Implicit in this model was the assumption that actinomycin binds to B-DNA, or a distorted B-DNA form. The possibility that actinomycin might bind to some other discretely different DNA conformational-state was not envisioned at that time.
A modification to this actinomycin-DNA binding model was subsequently proposed, which allows one to understand its mechanism of action (see Figure 2). This model is similar to the previous one; however, it predicts that actinomycin to bind to (what we have called) beta-DNA (i.e., not to B-DNA), this being a metastable and hyperflexible premelted form inferred from our wider crystallographic studies of planar drug molecules intercalated into a series of DNA-like and RNA-like self-complementary dinucleoside-monophosphates [12–14].
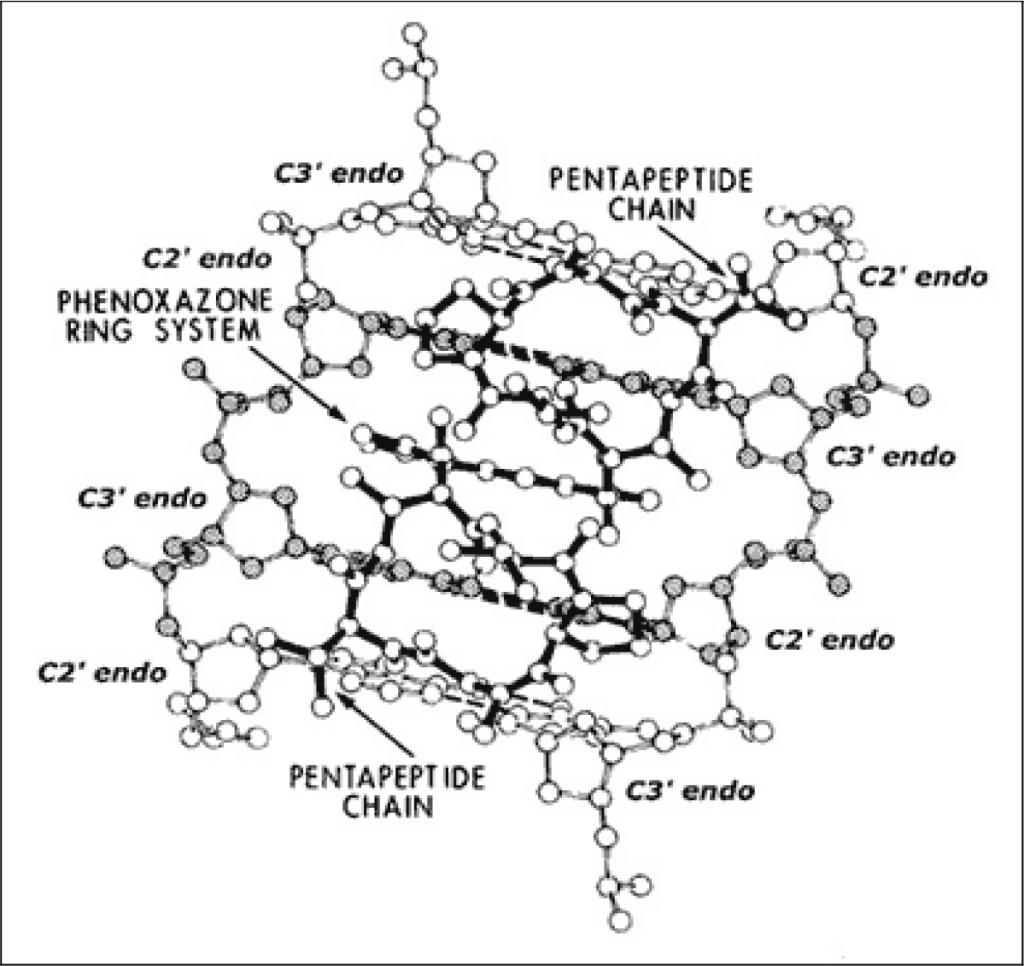
Figure 2. Actinomycin: beta-DNA binding model.
Figure 3 shows this same (extended) beta-DNA structure “pinned” by ethidium. The complex is an organized right-handed double helical structure in which the beta-structural element plus the intercalator form the asymmetric unit of the helix. This maximally elongated and unwound DNA duplex-structure, pinned by ethidium at saturating concentrations, readily explains the well-known observation of neighbor-exclusion intercalative drug-binding [15–17].
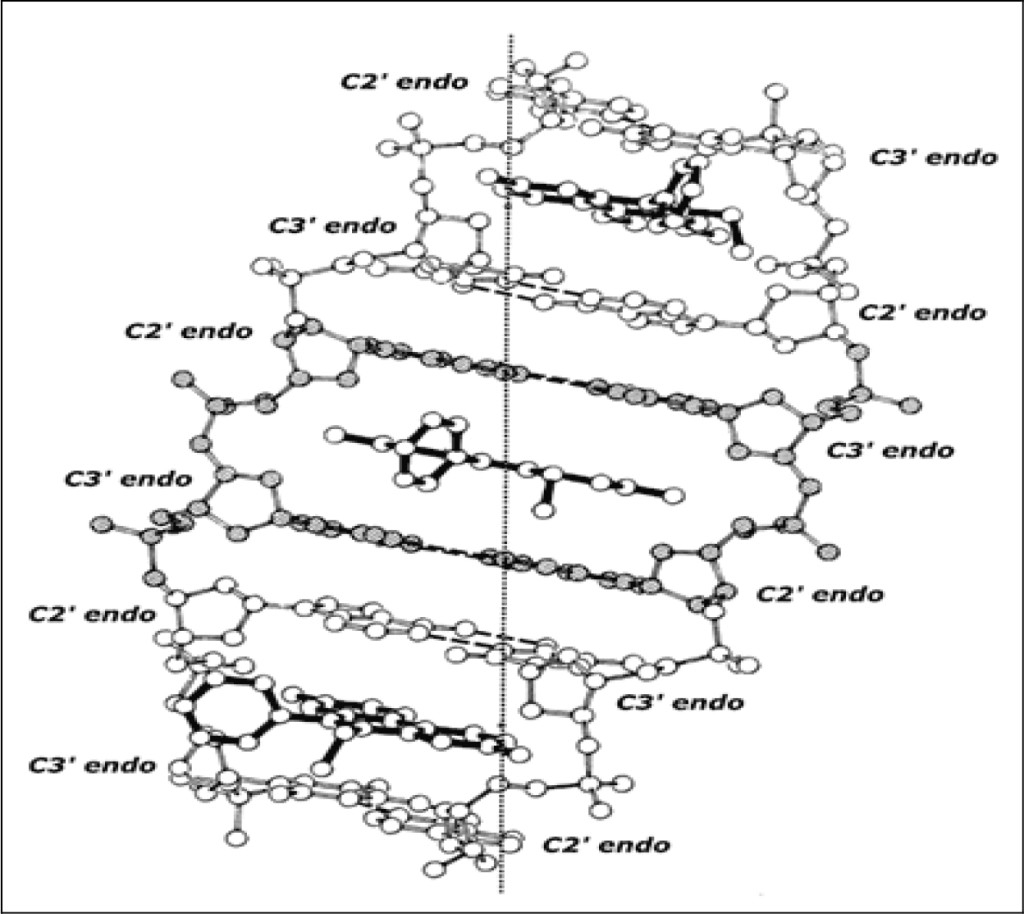
Figure 3. Ethidium: beta-DNA binding model
Mechanism of Action of Actinomycin D
I have next proposed beta-DNA to be an obligatory intermediate (i.e., a transition-state intermediate) in DNA-melting. This concept readily leads to understanding the mechanism of action of actinomycin D.
Figure 4 (a) shows an electron-micrograph of nucleolar 5-S ribosomal-RNA genes undergoing very active transcription in malignant Hela cells [18] and my interpretation of this process (b) — which indicates the mechanism of action of actinomycin D [19, 20].
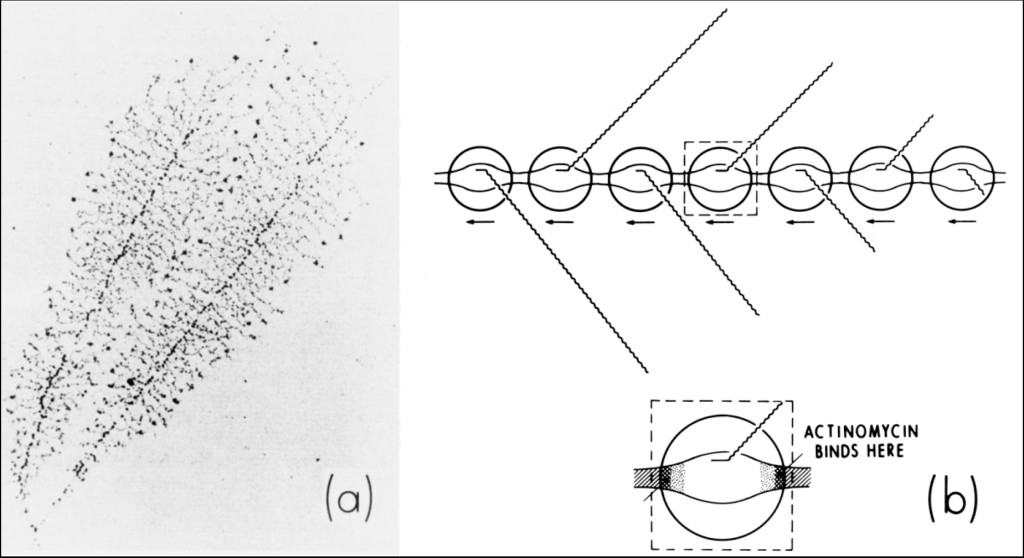
Figure 4. (a) Electron-micrograph of nucleolar 5-S ribosomal-RNA genes undergoing very active transcription within dividing amphibian oocytes [18]. (b) Interpretation of this photomicrograph showing how actinomycin might act to inhibit this process. Actinomycin binds to beta-DNA, a conformational intermediate that exists within the boundaries connecting double-stranded B-DNA with single-stranded DNA in the transcription complex. This immobilizes (i.e., “pins”) the complex, interfering with the elongation of growing RNA chains.
Actinomycin intercalates into beta-DNA found within the boundaries connecting double-stranded B-DNA with single-stranded DNA in the transcription-complex. This immobilizes (i.e., “pins”) the complex, interfering with the elongation of growing RNA-chains. In extremely active genes such as these, RNA polymerases lie in a close-packed arrangement along DNA. Interference with the movement of one polymerase by actinomycin is expected to inhibit the movement of other polymerases. This predicts nucleolar 5-S ribosomal RNS synthesis to be extremely sensitive to the presence of actinomycin [21–25].
Might actinomycin be expected to preferentially kill malignant cells?
Nucleoli within each nucleus in malignant cells (such as Hela-cells) are expected to contain large numbers of tandem repeats of 5-S ribosomal genes undergoing transcription. Since rapidly dividing malignant cells require increased numbers of ribosomes to carry out protein synthesis, they need many more tandem repeats of 5-S ribosomal genes per nucleoli than normal cells (or alternatively, it is possible that each nucleus within a malignant cell contains many more nucleoli than normal cells, the number of tandem repeats of 5-S ribosomal genes in each nucleolus remaining the same).
The presence of an effect such as this could allow actinomycin to preferentially kill malignant cells. For this reason, trace amounts of actinomycin given over extended periods of time can be expected to be a powerful anticancer chemotherapeutic regimen. Of course, it is first necessary to carry out the appropriate experiments in mice or in other related mammals before attempting clinical use.
References
- KIRK JM (1960) The mode of action of actinomycin D. Biochim Biophys Acta 42: 167–169.
- GOLDBERG IH, RABINOWITZ M (1962) Actionmycin D inhibition of deoxyribonucleic acid-dependent synthesis of ribonucleic acid. Science 136: 315–316.
- GOLDBERG IH, RABINOWITZ M, REICH E (1962) Basis of actinomycin action. I. DNA binding and inhibition of RNA-polymerase synthetic reactions by actinomycin. Proc Natl Acad Sci USA 48: 2094–2101.
- Reich E, Goldberg IH (1964) Actinomycin and nucleic acid function. Prog Nucleic Acid Res Mol Biol 3: 183–234.
- Sentenac A, Simon EJ, Fromageot P (1968) Initiation of chains by RNA polymerase and the effects of inhibitors studied by a direct filtration technique. Biochim Biophys Acta 161: 299–308.
- Perry RP (1963) Selective effects of actinomycin D on the intracellular distribution of RNA synthesis in tissue culture cells. Exp Cell Res 29: 400–406
- Goldberg, IH (1975) Handb Exp Pharmacol. 38: 582–592
- Sobell HM, Jain SC, Sakore TD, Nordman CE (1971) Stereochemistry of actinomycin–DNA binding. Nat New Biol 231: 200–205.
- Jain, SC, Sobell, HM (1972) Stereochemistry of actinomycin binding to DNA. I. Refinement and further structural details of the actinomycin-deoxyguanosine crystalline complex. J Mol Biol 68: 1–20
- Sobell HM, Jain SC (1972) Stereochemistry of actinomycin binding to DNA. II. Detailed molecular model of actinomycin-DNA complex and its implications. J Mol Biol 68: 21–34.
- Sobell HM (1974) How actinomycin binds to DNA. Sci Am 231: 82–91.
- Sobell HM (1985) Actinomycin and DNA transcription. Proc Natl Acad Sci USA 82: 5328–5331.
- Sobell, HM (2009) Premeltons in DNA, Explanatory Publications, Lake Luzerne, NY ISBN 978-0-615-33828-6
- Sobell, HM (2013) Organization of DNA in Chromatin, Explanatory Publications, Lake Luzerne, NY ISBN 978-0-692-01974-0
- Crothers, DM (1968) Calculation of binding isotherms for heterogeous polymers. Biopolymers 6: 575–583
- Wells RD, Larson JE (1970) Studies on the binding of actinomycin D to DNA and DNA model polymers. J Mol Biol 49: 319–342.
- Bond PJ, Langridge R, Jennette KW, Lippard SJ (1975) X-ray fiber diffraction evidence for neighbor exclusion binding of a platinum metallointercalation reagent to DNA. Proc Natl Acad Sci USA 72: 4825–4829.
- Miller OL Jr, Beatty BR (1969) Visualization of nucleolar genes. Science 164: 955–957.
- Sobell HM1 (2016) Premeltons in DNA. J Struct Funct Genomics 17: 17–31.
- Sobell, HM (2016) How actinomycin binds to DNA and exerts its mechanism of action. J Struct Funct Genomics
- Sobell, HM (2017) World Journal of Pharmaceutical Research 6: 78–84
- Sobell, HM (2018) International Journal of Biopharmaceutical Sciences Boffin Access UK Premeltons in DNA (in press)
- Sobell, HM (2018) International Journal of Biopharmaceutical Sciences Boffin Access UK ` Organization of DNA in Chromatin (in press)
- Leroy Liu and James Wang have provided a key insight into the nature of DNA supercoiling accompanying transcription that has shed additional light on this question. They have theorized that – in the presence of significant resistance to the rotational motion of the RNA polymerase and its nascent RNA chain around DNA during transcription – the advancing polymerase generates positive superhelicity in the DNA template ahead of it, and negative superhelicity behind it. In nucleolar genes, where there may as many as 200 RNA polymerases moving down the DNA template while synthesizing growing ribosomal RNA-chains, positive and negative superhelical DNA regions between them annihilate one-another, causing adjacent chains to bond-together to form “trains” of transcription complexes, these now moving synchronously along DNA. If this were the case, then the binding by one actinomycin molecule is sufficient to stop the entire “transcription-train” from moving along DNA.
- Liu LF1, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84: 7024–7027.