Abstract
P. tricornutum has been found in several locations worldwide. This study first isolated a P. tricornutum strain from the east coast of Australia. The newly isolated strain grew well at salinity levels from 25 ppt to 45 ppt. However, it could not survive when the temperature was higher than 30°C. Ammonia was toxic to this species when the concentration was higher than 2 mM, and ammonium can be used as an alternative nitrogen source for this species. The main fatty acids are C16:0, C16:1 and C20:5 (eicosapentaenoic acid; EPA), together accounting for 85% of total fatty acids, and the EPA content was about 4% per dry weight. After nutrient starvation, the total fatty acid accumulated in the newly isolated strain at 25 ppt (by 110%) and 35 ppt (by 76%) salinity levels, as well as EPA content per dry biomass. P. tricornutum has the potential for the EPA production.
Keywords
Phaeodactylum tricornutum, Salinity, Temperature, Eicosapentaenoic acid
Introduction
With the increasing emissions into the atmosphere, the concentration of CO2 increased not only in the air but also in the ocean [1]. The greenhouse effect may affect the nutrient content in the ocean as well as the distribution of marine diatoms. The cell size of P. tricornutum is significantly smaller by approximately 15% under N-limited conditions. However, with simulated increased CO2 concentrations (expected by the end of this century), the growth rate was not significantly increased [1]. Brisbane is located in the southeast corner of Queensland, Australia, and has a humid subtropical climate. The minimum mean temperature is 16.6°C and the maximum mean temperature is 26.6°C. East coast sea water temperatures peak in the range of 26°C to 28°C around early February and the lowest in about mid August, in the range 20°C to 22°C. P. tricornutum has been found in several places around the world, typically in coastal areas with wide fluctuations in salinity [2]. Therefore, it is reasonable to consider that the local water system may harbour this species. This part of the research isolated a local strain of P. tricornutum and discovered new properties from it, such as ammonia tolerance. Growth and EPA content comparisons were made between local strains and control strains (Tasmania originated strains).
Materials and Methods
Sample Collection and DNA Extraction
Water samples were collected from the Brisbane River, Gold Coast, Moreton Bay, and Yamba. All samples were stored in 500 mL sealed bottles. A light microscope (OLYMPUS CX21LEDFS1) was used to identify the morphtypes of the cells. After that, a 100 µL sample was used for the DNA extraction by using a DNA extraction kit (DNeasy Plant Mini Kit) according to the manufacturer’s instruction. The extracted DNA was stored at -20°C. The control strain used in this study for benchmarking is P. tricornutum CS-29/8, which originates in Tasmania and was obtained from the Commonwealth Scientific and Industrial Research Organisation (CSIRO) and stored in the Queensland Microalgae Culture Collection.
Specific Primers Design
Specific primers were designed for P. tricornutum. The length of the primers should be around 20 bases, and the G-C composition should be 40%-60%. Before submitting requests for synthesis, the designed primers were checked for self-annealing and potential primer dimer formation. In addition, if homologies to non-target regions higher than 70% were found, those primers were not used. Table 1 shows the primers used in this study. The primers were synthesised by Integrated DNA Technologies, Inc.
Table 1: Primers used in this study.
|
Name |
Sequence 5’-3’ |
Tm | GC content |
length |
|
e-cls1-F |
TCGGCAGTTACAATCCCCAC | 57.4°C | 55.0% |
20 |
|
e-cls1-R |
AATGCCCACGCCAAGAGTAA | 57.1°C | 50.0% |
20 |
|
5.8s-F |
TCGGCGTCTTTTTACCACGA | 56.8°C | 50.0% |
20 |
|
5.8s-R |
GTATCGCATTTCGCTGCGTT | 56.4°C | 50.0% |
20 |
PCR and Electrophoresis
After DNA extractions, PCR assays were performed. The total reaction volume for PCR was 25 µL, which contains 1 µL extracted DNA templates, 5 µL 5xBuffer (Mg2+-free), 0.5 µL dNTPs (10 mM), 0.5 µL MgSO4 (100 mM), 0.5 µL primers (10 µM, forward and reverse), 0.125 µL Taq DNA polymerase, and 16.875 µL PCR grade water. The program for PCR was set as follows: stage 1, 95°C for 5 min; stage 2, 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min; stage 3, 72°C for 10 min and hold on 10°C. For gel electrophoresis, 5 µL of PCR products were mixed with 1 µL DNA Gel Loading Dye (6X). Then the mixtures were injected into the slots of an agarose gel (2% TAE buffer, with 1 drop of ethidium bromide). The current was set to 110 mA (BioRAD PowerPac300). After 30 min, the gel was placed onto the gel reader (UVItec BIS-26.LM).
Single Colony Isolation
For this part of the research, a 1.5% agar plate was used to cultivate water samples, which contained F/2 medium with 35 ppt sea salts. 100 µL samples were spread onto the surface of the agar plate. The cells grew slowly in the solid medium. After the formation of a single colony (approximately 30 days), a sterilised loop was used to pick up the single colony and then suspended in liquid medium.
Scale Up and Optimise Growth Conditions
After the newly isolated P. tricornutum strains had been successfully grown in the liquid medium, growth conditions were measured under different temperatures and salinities, and a comparison was made between newly isolated strains and previously stored strains. For the temperature test, temperature pads (heat mats) were used and set to 25°C, 30°C, and 35°C. The growth conditions were measured using the optical density (OD) value at 750 nm. A spectrophotometer (UV-1800, Shimadzu, Japan) was used in this study. For the salinity test, 25 ppt, 35 ppt, and 45 ppt sea salt levels were used to cultivate both strains (distilled water mixed with commercial sea salt). The ammonia tolerance was also tested in this study: different concentrations of ammonium sulfate were prepared in the culture. The pH was adjusted to 8 and 10 by using 1 N NaOH and HCl. Nitrate and phosphate concentrations were tested every day during the experiment by using API Nutrient testing kits according to the user’s manual. Algae were cultivated in 500 mL flasks in this study, and grown in a light-controlled room with 16 h light and 8 h dark cycle. The light intensity was set to 100 μmol m−2 s−1. Filtered (0.45 µm) air was pumped into the flasks with 137.2 kPa.
Fatty Acid Analysis
For fatty acids analysis, a previously reported method was used [3,4].
Results and Discussion
Samples Collection and DNA Extraction
Samples were collected from different areas around Brisbane. They were from Brisbane River (in the Brisbane city), Gold Coast (south-east of Brisbane), Moreton Bay (north of Brisbane), and Yamba (further south of Brisbane). Figure 1 shows the microscopic images of all water samples. All samples contained microorganisms, however, different samples had different dominant microbes. The fusiform microbes account for the majority in the Yamba sample (bottom right). P. tricornutum shows the fusiform shape when grown in liquid medium, however, oval cells occupy the bulk part when grown in solid medium [5]. Therefore, it is reasonable to suspect that the Yamba sample contains P. tricornutum, however, further DNA evidence is required.
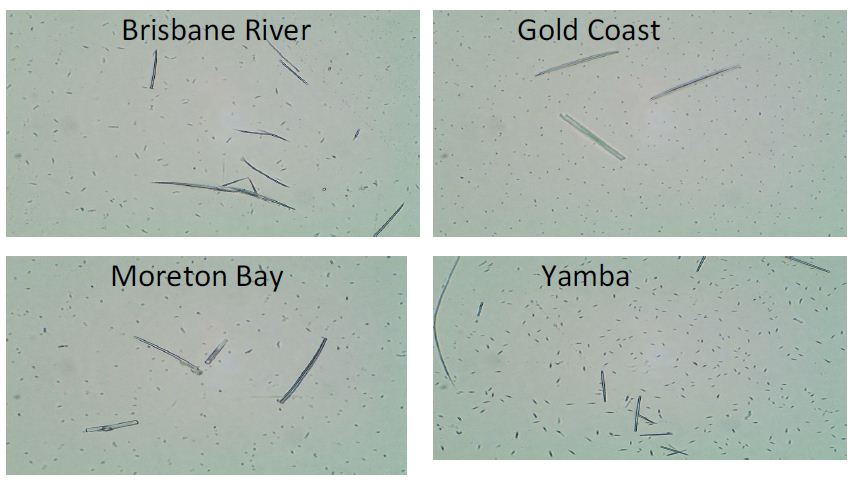
Figure 1: Microscopic imaging of samples from Brisbane River, Gold Coast, Moreton Bay, and Yamba with 100x magnification.
PCR Results
Figure 2A shows the electrophoresis result of using 5.8S rDNA primers for PCR using DNA from the water samples as template. The 5.8S ribosomal RNA gene is widely found in microalgal species and regulates the synthesis of the large subunit of ribosomes. Samples from the Gold Coast, Moreton Bay, and Yamba had a positive result, which indicated that all these three samples contained detectable microalgal species. However, samples from Brisbane River showed no bands, which indicates that the microalgal content might be lower than the detection limit. In order to further confirm whether the water samples contain P. tricornutum, specific primers (e-cls1) were used to identity this species. P. tricornutum eukaryotic-type cardiolipin synthase 1 (e-cls-1) gene is a specific gene of this species [5]. When subjecting the gene sequence to the Basic Local Alignment Search Tool (BLAST) it was confirmed that only P. tricornutum contains this gene in Genbank data. Figure 2B shows the electrophoresis result of using e-cls1 primers for PCR. Just as expected, Yamba samples (line 7 & 8) gave positive results, which indicates that the P. tricornutum content from Yamba is higher than that from other samples. The sample from Yamba was hence selected for further isolation.
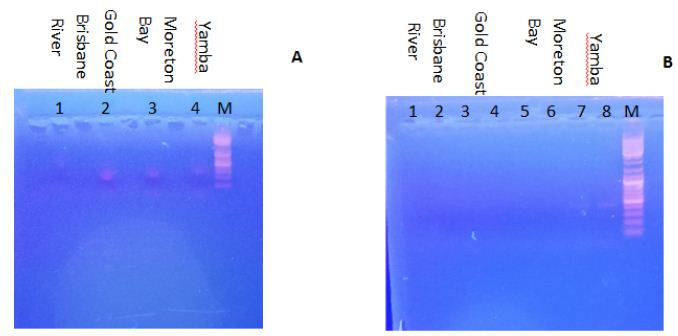
Figure 2: Electrophoresis results. A. The use of 5.8S rDNA primers. Lines 1, 2, 3 and 4 indicate the different DNA templates from water samples. Line 1 is from the Brisbane River; Line 2 is from the Gold Coast; Line 3 is from Moreton Bay; Line 4 is from Yamba; Line M is the marker; B. The use of using e-cls1 primers. Line 1 to 8 indicate the different DNA templates from different water samples. Line 1 and 2 are from the Brisbane River; Line 3 and 4 are from the Gold Coast; Line 5 and 6 are from Moreton Bay; Line 7 and 8 are from Yamba; Line M is the marker.
Single Colony Isolation
Solid medium and semi-solid medium could be used for the isolation of microalgae, and it is easy to pick up individual colonies [6]. The water sample from Yamba was spread onto the surface of an agar plate (solid medium). The cells grew slowly in the solid medium. In solid medium, the main shape of the cells was oval. However, in liquid medium, the majority of the shapes was fusiform. Only P. tricornutum possesses this property. The ovoid cell walls contain silicified frustules, and are five times stiffer than the other two shapes. And the maximal growth rate of fusiform cells is 1.4 times higher than oval cells [5]. After approximately 30 days’ cultivation, single colonies were formed (Figure 3A). Five different colonies were selected to continue growth in liquid medium (F/2 nutrients with 35 ppt sea salt). After another 15 days’ cultivation, two flasks showed a brown colour, which indicated successful growth of P. tricornutum (Figure 3B). This (4th) flask had more biomass and displayed dark brown colour, which suggested that P. tricornutum was successfully isolated in this flask. Figure 3C shows a microscopic photo of P. tricornutum in the 4th flask after 15 days of cultivation. The shape of the cells underwent a transition from oval (in the solid medium) to fusiform (in the liquid medium), which further confirmed that P. tricornutum was successfully isolated. The newly-isolated strain was named “Yamb” in this study.

Figure 3: Colonies isolation. A. Single colonies on solid medium after 30 days of cultivation; B. 5 different colonies grew in liquid medium after 15 days; C. Microscopic picture of a cell in the 4th flask with 400x magnification.
Growth Test
After this local P. tricornutum strain had been successfully isolated, a comparison was made to the previously stored strain CS-29/8 under different growth environments. Temperature is an important factor that can strongly affect the growth of this species. Jiang [7] tested the growth conditions of P. tricornutum at different temperatures (10, 15, 20, 25 and 30°C). They found that the optimum temperature for P. tricornutum growth was 20°C, and it grew slower at higher or lower temperatures, and it hardly showed any growth at 30°C. Bernard [8] constructed a model for the prediction of the growth rate of P. tricornutum, according to the published datasets. In this model, the optimal temperature for growth of P. tricornutum is around 23°C. According to the model, the growth rate decreases sharply at higher temperatures, however, it decreases gradually towards lower temperatures. In the present study, 25°C, 30°C, and 35°C were used to test the growth conditions of the newly isolated strain (Yamb), as well as the previously obtained strain CS-29/8 (as a control), as it was hypothesised (based on its original habitat) that the new strain may display better growth tolerance at higher temperatures. Figure 4A shows the growth conditions of both strains under different temperatures. However, both strains could not survive at 30°C or higher, which is consistent with former results [8]. This phenomenon could be explained by the decrease of photosynthesis rate as well as the carbon assimilation when the temperature is higher than 25°C [9]. Moreover, the relative expression level of small heat-shock protein (shsp) gene is 566 folds higher under thermal stress [10], which may further reduce the growth of this species. Although isolated from a location with warm climate, the local strain could still not survive when the temperature was 30°C or higher, which suggested that P. tricornutum cannot grow in open ponds during the hottest months of the year (From December to February). As for the salinity test, the growth-permitting salinity of P. tricornutum ranges broadly from 5 ppt to 70 ppt [2]. There were no significant differences in growth conditions of salinity levels ranging from 25 ppt to 45 ppt for both strains (Figure 4B). 35 ppt (sea water salinity) was the best growth environment for this species, and was selected to use throughout this study.
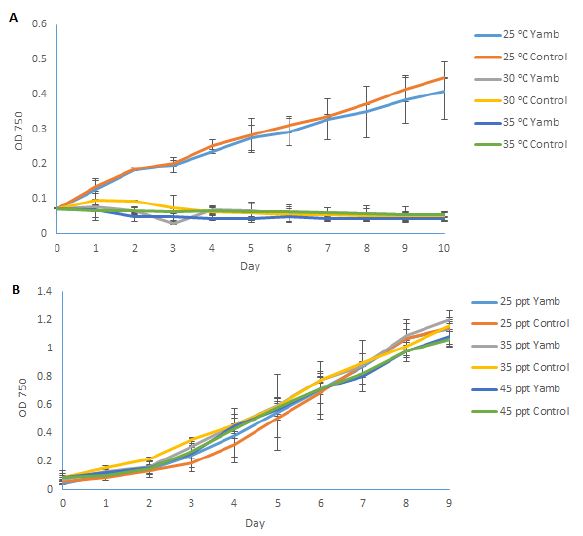
Figure 4: Growth conditions of P. tricornutum under different temperatures (A); and salinities (B). Yamb: newly isolated strain; Control: Tasmania-originating strain CS-29/8. Shown are mean values ± SD of three separately-grown cultures, each.
Ammonia (NH3) can be used as an additional nitrogen source for microalgae. When dissolved in water, there is an equilibration between ammonia and ammonium (NH4+):
NH3 + H2O↔NH4+ + OH–
If the pH decreases, the equilibration moves to the right side, and more ammonia molecules are converted into ammonium. On the contrary, if the pH increases, the equilibration moves to the left side, and more ammonia molecules are in the solution. It has been reported that, when ammonia was used as the main nitrogen source without adjusting the pH, the growth rate P. tricornutum was very low, about 10% of cell numbers per millilitre compared to a nitrate-based culture, and the pH dropped to below 5 [11]. If the pH was adjusted to 8.2, the higher concentration of ammonia could also inhibit the growth of P. tricornutum. It has been reported that 545 gene transcripts altered in P. tricornutum under ammonia treatment [12]. In the present study, different concentrations of ammonium sulfate were used, and the pH was adjusted to 8 and 10. Figure 5A shows the growth conditions of P. tricornutum in different ammonia concentrations at pH 8. Growth was certainly inhibited by the increasing concentrations of ammonia, which in accordance with previous research [12]. And the growth conditions decreased sharply when the ammonia concentration was higher than 2 mM after 4 days of cultivation. Figure 5B shows the growth curves of P. tricornutum in different ammonia concentrations at pH 10. The cultures became cloudy when the pH was adjusted to 10, due to the precipitation of Ca(OH)2 in the culture. These results also indicate that ammonia was toxic to P. tricornutum when the concentration was higher than 2 mM, and that ammonium can be used as an alternative nitrogen source to grow this species. Knowledge of the ammonia tolerance of P. tricornutum is also important when using ammonia to control predators in the culture.
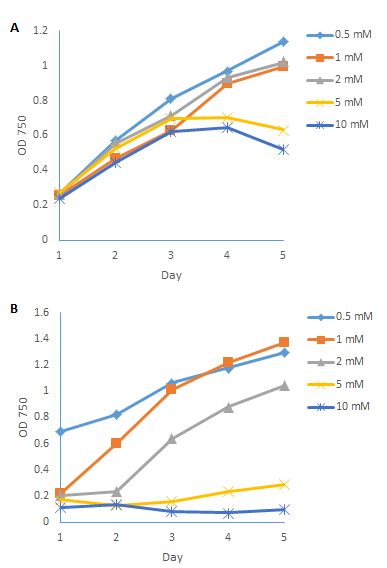
Figure 5: Growth conditions of P. tricornutum in different ammonia concentrations under pH 8 (A); and pH 10 (B).
Fatty Acids Analysis
Different strains of P. tricornutum may have different lipid profiles, as well as total fatty acid compositions. In this study, fatty acid profiles were analysed. Figure 6A shows the fatty acid content per dry biomass of both strains at different salinity levels on day 5 and day 8 after inoculation. The nitrate ran out on day 5. After this, the cells went into the stationary growth phase, and day 8 was in the early stationary growth phase. EPA contents increased from day 5 to day 8 under 25 ppt salinity level for both strains, as well as the Yamb strain under 35 ppt. However, there were no significant differences in the control strain CS-29/8 from day 5 to day 8 under the 35 ppt salinity level. As for 45 ppt, the EPA content decreased from day 5 to day 8 for both strains. The total fatty acid content per dry biomass showed a similar trend for both strains (Figure 6B). After nutrient starvation, the total fatty acid contents per dry biomass increased in the Yamb strain under 25 ppt (by 110%) and 35 ppt salinity (by 76%) levels. The fatty acid composition of both strains under different salinity levels displayed no significant difference on day 5 and on day 8 (Figure 6C). However, a decrease of EPA percentage was observed from day 5 to day 8 in both strains. Accordingly, the proportion of saturated fatty acids and monounsaturated fatty acids (C16:0 and C16:1) increased. Taken together, this indicates that, after nutrient starvation, both strains could accumulate EPA and total fatty acids under a low salinity level (25 ppt). However, the contents of EPA and total fatty acid per dry biomass decreased under a higher salinity level (45 ppt). The percentage of EPA of total fatty acids decreased from day 5 to day 8; however, the proportion of saturated fatty acids and monounsaturated fatty acids (C16:0 and C16:1) increased in both strains. There were no significant differences in the EPA content between both strains from 25 ppt to 45 ppt. C16:0, C16:1, and C20:5 (EPA) are the main fatty acids for this species, and together account for approximately 85% of the total fatty acids. The acyl-CoA pool composition of P. tricornutum also indicated that C16:0, C16:1, and EPA were the most abundant fatty acids (Hamilton et al., 2014). The EPA content per dry biomass could reach 5% in the present experiment, which indicates that P. tricornutum is a promising candidate that could be used for EPA production.
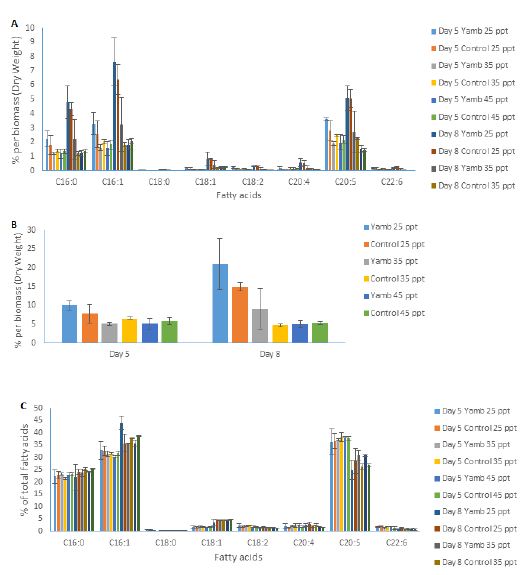
Figure 6: Fatty acid profile of P. tricornutum at different salinity levels on day 5 and day 8 after inoculation. Graphs show A, Fatty acid contents per dry biomass; B, Total fatty acid contents per dry biomass; C, Fatty acid composition. Shown are mean values ± SD of three separately-grown cultures.
Conclusion
A local P. tricornutum strain had been successfully isolated from the east coast of Australia and named Yamb in this study. Growth comparisons between P. tricornutum Yamb and P. tricornutum CS-29/8 showed that there are no significant differences. Both strains cannot survive when the temperature is higher than 30°C. It seems that P. tricornutum has a better living strategy when exposed to different salinity levels. Both strains grew well from 25 ppt sea salt level to 45 ppt sea salt levels. Ammonium could be used as an additional nitrogen source for this species, and it could facilitate the growth of P. tricornutum when the pH is at 8. However, ammonia is toxic to this species when the concentration is higher than 2 mM at pH 10. C16:0, C16:1, and C20:5 (EPA) are the main fatty acids for this species, together accounting for about 85% of the total fatty acids. The EPA content per dry biomass could reach 5% in both strains, which makes P. tricornutum a potential source for EPA production.
Acknowledgement
This work was supported by a Cooperative Research Centre Project (CRC-P50438) jointly funded by the Australian Government, Qponics Limited, Nutrition Care Pharmaceuticals and The University of Queensland, and an Advance Queensland Biofutures Commercialisation Program (AQBCP00516-17RD1) jointly funded by the Queensland Government, Woods Grain Pty Ltd and The University of Queensland.
References
- Li W, Gao K, Beardall J (2012) Interactive Effects of Ocean Acidification and Nitrogen-Limitation on the Diatom Phaeodactylum tricornutum. PLoS ONE
- Cui Y, Thomas-Hall SR, Schenk PM (2019) Phaeodactylum tricornutum microalgae as a rich source of omega-3 oil: Progress in lipid induction techniques towards industry adoption. Food Chemistry
- CuiY, Thomas-Hall SR, Chua ET, Schenk PM (2020) Development of high-level omega-3 eicosapentaenoic acid (EPA) production from Phaeodactylum tricornutum. Journal of Phycology.
- Sabir JSM, Theriot EC, Manning SR, Almalki AL, Khiyami MA, et al. (2018) Phylogenetic analysis and a review of the history of the accidental phytoplankter, Phaeodactylum tricornutum Bohlin (Bacillariophyta). PLOS ONE
- Tian HF, Feng JM, Wen JF (2012) The evolution of cardiolipin biosynthesis and maturation pathways and its implications for the evolution of eukaryotes. BMC Evolutionary Biology [crossref]
- Cho JY, Choi JS, Kong IS, Park SI, Kerr, et al. (2002) A procedure for axenic isolation of the marine microalga Isochrysis galbana from heavily contaminated mass cultures. Journal of Applied Phycology 14: 385-390.
- Jiang H, Gao K (2004) Effects of lowering temperature during culture on the production of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum (bacillariophyceae). Journal of Phycology 40: 651-654.
- Bernard O, Rémond B (2012) Validation of a simple model accounting for light and temperature effect on microalgal growth. Bioresource Technology 123: 520-527.
- Li KW, Morris I (1982) Temperature adaptation in Phaeodactylum tricornutum Bohlin: Photosynthetic rate compensation and capacity. Journal of Experimental Marine Biology and Ecology 58: 135-150.
- Egue F, Chenais B, Tastard E, Marchand J, Hiard S, et al. (2019) Expression of the retrotransposons Surcouf and Blackbeard in the marine diatom Phaeodactylum tricornutum under thermal stress. Phycologia 54: 617-627.
- Fidalgo JP, Cid A, Abalde J, Herrero C (1995) Culture of the marine diatom Phaeodactylum tricornutum with different nitrogen sources: Growth, nutrient conversion and biochemical composition. BioI. Mar 36: 165-173.
- Osborn HL, Hook SE (2013) Using transcriptomic profiles in the diatom Phaeodactylum tricornutum to identify and prioritize stressors. Aquatic Toxicology 138-139. [crossref]