Abstract
This paper described a hypothesis about phenomenon those may have occurred early in the evolution of life or pre-life to reveal how every creature of the Earth has strong preference for specific chirality. Thread of helical molecule cannot be replicated by using a casting mold at once. Replication of creature has been carried out by DNA. The interactions among molecules of DNA are able to estimate from stereo structure of DNA. There are a D-typed chiral strand and a L-typed chiral strand linked by bases in a DNA. Those strands are generated simultaneously in the vicinity, but two will not coalesce due to the different typed chirality of the helix. The paired strands of DNA are able to become a representative of a series of amino acids for a protein i.e., DNA is able to memorize the information of replication of a protein. If amino acid molecules will be bonded with other amino acid at random timing, the alignment of amino acids is randomized in the protein. In such case, the replication is not accurate, and it will be eliminated by natural selection. Current amino acids alignment is stored by using genetic code in DNA, and replication of protein is constructed by using genetic codes sequences sequentially. In the double helix of DNA, there are leading strands and lagging strands, and the two strands proceed in rotationally opposite directions to find matched base pairs, the protein is formed when two genetic codes are in a key-keyhole relationship. The processing is carried out by amino acid units. The nucleobase ciphers of codon and anticodon represent amino acids at matching processing. The chirality of biomolecules takes one of the most important roles in the activities of creature in the Earth.
Keywords
Protein, Chirality, Helix, DNA, Leading strand, Lagging strand, mRNA, tRNA
Introduction
Although we have much knowledge in the field of molecular biology, the reason for biology’s strong preference for specific chirality of amino acids, sugars, and other molecules remains unanswered question in the field of life research [1]. S. Karasawa described that origin of life can be explained by intermolecular interactions among molecules via a helical structure of water [2]. After that, the study was advanced by focusing helical of DNA. This paper is the results of such studies. C. R. Cantor and P. R. Schimmel presented from mathematical requirements that asymmetric molecules can only have a helical structure as a polymer structure [3]. L. E. MacKenzie and P. Stachelek reported that the chirality twisted molecular structures will interlock and bind only if their chirality allows them. The chirality of a molecule interacts selectively with other molecules [4]. A. Shimada proposed that thing that separates life from non-life is enantioselectivity which escape from racemization [5]. N. Nemoto described that the chirality of biopolymers might depend on thesynthetic process of the biopolymer units [6]. The formation of biochemical molecules depends on the surrounding environment, and the homochirality of creature was established though natural selection that allowed to survive the creature fit for to live. K. Tamura pointed that biological homochirality could have been determined through the process of coevolution between nucleotides and amino acids. And he reported that aminoacylation of tRNA could be the key step in the origin of amino acid homochirality and once L-amino acids had been selected, the elongation of L-amino acids by the ribosome would have synthesized proteins composed of L-amino acids [7,8]. Current building blocks of amino acids are left-handed (L-type) chiral molecules, and ribose is right-handed chiralty (D-type). L-type amino acids never mingle with nucleic acids made of D-type sugars in a cell. K. Tamura and P. Schimmel showed that an RNA minihelix was aminoacylate by an aminoacyl phosphate D-oligonucleotide with a clear preference for L- as opposed to D-amino acids [9]. As the result of evolution, current ragging processing of D-type lagging strand in replication of DNA includes stops, turns, and synthesizes intermittently as shown in Okazaki fragment [10]. The double helix of DNA cannot form without selectivity of the chirality. The chirality of molecules supports self-assembly of molecules and formation of protein by using complex enzymes those are made by proteins. However, the replication of DNA has not fully discussed from the viewpoint of interaction among helical structures up to now. S. Karasawa described that if amino acid molecules are related to mRNA physically and amino acids bind at random timing to mRNA, accurate replication is impossible [2]. Current alignment of amino acids for formation of protein is realized by the genetic codes as merely representatives of amino acids. When codon and anticodon are in a key-keyhole relationship in the matching process, the amino acid is settled correct position, and it makes possible to reproduce the accurate protein.
The Environment for Formation of Cell Membrane
Movements of Liquid Water Molecules
The biochemical reactions are carried out in environment of liquid water. Even when ice melts into liquid water, there exist many of hydrogen bonds. The enthalpy of hydrogen bond of water is about 21 kJ/mol, and the change of the thermal energy from ice to water is 6.0 kJ/mol. There exist more than 90 % of hydrogen bonds among molecules in liquid water. The hydrogen bonds will generate many clusters in the liquid water. Since the tetrahedral molecule of water has an electric polarization, and it assists to form a cluster of helical structure similar to α-quartz that is minimum size with minimum energy in stereo structure made of asymmetric tetrahedrons [11]. The structure of liquid water is constantly being generated and extinguished, and its average lifespan is about 10-12 seconds. Helical structure of liquid water is similar to lattice structure of α-quartz, and it is minimum size with minimum energy in lattice structure of tetrahedrons The phase transition of quartz from β to αcan be explain as the shrink of size by rotation of unit SiO4 alternately around the electrical axis to change the symmetry from hexagonal to trigonal, and the tetrahedral unit projected to the X-Y plane from square to trapezoidal. The unit structure of helix is aligned with the central shafts stand on a planar boundary vertically and it has three-rotational symmetrical electrical axes. When the pair of hydrogen atoms located at the ends of the long and short sides vibrates around electrical axis, the up and down movements of the molecular pair of the inner vertices of tetrahedron and vertices of the outer side moves in the opposite direction. The movements of asymmetrical tetrahedrons make possible to form helical structure of DNA. Since helix structure is minimum size with minimum energy, it has tendency to expand more wider area. So, the clusters of helixes are formed. Even though the lifetime of the cluster is very short, the coupling among the helical area it yields rapid biochemical interactions. As evidence of connection by the helical state of water, linked bubbles are observed at thawing ice of carbonated water.
Catalytic Effects due to Helical Structure of Water Molecules
Proteins are made up of amino acids. On the other hand, nucleic acids are made up of nucleotides. Proteins and nucleic acids are different types of biomolecules those have different functions in the body. The control function is realized by different the chirality of molecules should be controlled and that of controller. Waves on the sea surface are happening all the time. Fluctuations of pressure in water causes the helical structure of water repeatedly expands and contracts via rotation of tetrahedron around each electric arises. Such repeated expansion and contraction of helical molecules will contribute to activities of a life. When a filamentous series of L-type of amino acids invades into the holes of helical structure, L-type structured of water will be long lifetime owing to the same chirality. The cluster of helix makes couplings with neighboring similar helical structures. The linkage takes role of a catalyst in the wet process. There are two types of helical molecular movements of liquid water as shown in Figure 1.
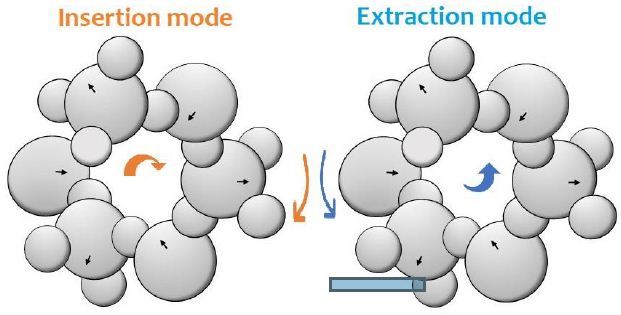
Figure 1: Helical structure of liquid water. Here, electric polarization in a water molecule is expressed by arrows. An oxygen atom is expressed by large circle, and a hydrogen atom is expressed by small circle.
Largely Linked Helical Movements of Molecules in a Membrane
Tamura reported that a clear preference for L-amino acids as opposed to D-amino acids was noted in the efficient nonenzymatic aminoacylation of an RNA minihelix (progenitor of the modern tRNA) by an aminoacyl phosphate oligonucleotide [7]. There exists linkage of L-type of helical movement caused by aligned L-type molecules. Interlinks among helical movements will induce large helical movements. The exist of large systematic helical movements will support to form large helical arrangement of molecules such as DNA. L-type of helical movements in the vicinity of L-type amino acid able to assist to synthesize L-typed helical filamentous molecules of protein. Current DNA is consisted of enormous number of molecules. When DNA had been first created, every element that makes up the mechanism of DNA must be incorporated at the same time. The first DNA had formed through interactions among molecules in the vicinity of plural organized molecules. Since asymmetric tetrahedral molecules can only have a helical structure as a polymer structure [3] and D-type of sugars exist in the cell, we can assume that D-type of helical movements on the surface of cell membrane able to assist to synthesize D-typed helical filamentous molecules of sugars. The rotational thermal oscillations of helical structures of asymmetric molecules will induce surrounding thermal motions. It is considered that when two kinds of chiral molecules rotate, the opposite directional rotation may occur by different chiral molecules by a gear mechanism as shown in Figure 2.
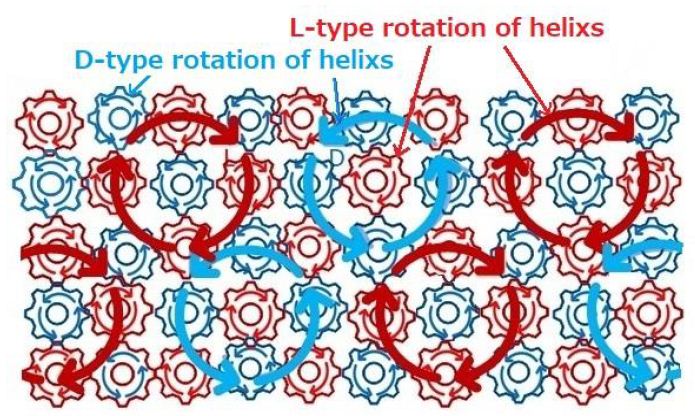
Figure 2: The illustration shows an image of only the mechanism that D-type of helical movements are induced by interactions among L-type of helical molecules. There exist large systematic helical movements.
Evolution of Cell Membrane
Origin of Main Component in Cell Membrane
The first life forms were born depending on the environment around them. The current cell membrane contains hydrophobic long chain of C16H34 or C18H38 as a main component. These molecules would have been in liquid state in the early Earth because these molecules are in liquid state between about from 20°C to 300°C. Since those molecules are hydrophobic long chains and the specific gravity are lighter than water, those molecules stay for long time as an oil film on the surface of water. Such hydrocarbon molecules had been synthesized from CO2 in the early atmosphere of the Earth due to the collision of the H+ of the solar wind. The smaller hydrocarbon molecules produced in the upper sky remain in the atmosphere and undergo repeated synthesis reactions [12]. In helical structure of liquid water, there are vacant shafts surrounded by a tetrahedron of water molecules as shown in Figure 1. The vacant shafts would have up-taken a C16H34 or C18H38, and those molecules aligned vertically on the water surface.
The processes of evolution of cell membrane are as follows.
- Fatty acids produced by oxidation of end terminal of hydrocarbons stayed in the hydrophobic layer.
- The membrane with linked head of fatty acids by glycerol laterally became robust.
- Long-chains polymeric carbohydrates hydrates (Cx(H2O)y) such as sugars tended to adhere on the hydrophobic layer.
- Helicase unwinds DNA, and two strands are used for replication of DNA.
- New bases are added to the complementary parental strands.
- Leading strand is made continuously, while lagging strands is made by pieces by piece.
- When nucleotides (bases) are matched, two of double helices are synthesized.
- Lee C, Jessica MW, Laura ER, Rachel YS, Laura MB, et al. (2022) Chirality in organic and mineral systems: a review of reactivity and alteration processes relevant to prebiotic chemistry and life detection missions. Symmetry 14.
- Karasawa S (2023) Origin of life in the water of the Earth. Geology, Earth & Marine Sciences 5: 1-7.
- Cantor CR, Schimmel PR ((1980)) Biophysical Chemistry: the conformation of biological macromolecules; P7, W H Freeman & Co., USA.
- MacKenzie LE, Stachelek P (2021) The twists and turns of chiral chemistry. Nature Chemistry 13: 521-522.
- Shimada A (2016) Thing that separate life from non-life – it’s enantioselectivity, Special Feature: The 41st Symposium, Viva Origino, 44, 3.
- Nemoto N (2016) A chirality of biopolymer from the perspective of evolutionary molecular engineering, Special Feature: The 41st Symposium, Viva Origino, 44, 5.
- Tamura K (2008) Origin of amino acid homochirality: relationship with the RNA world and origin of tRNA aminoacylation. Biosystems 92: 91-98.
- Tamura K (2016) Interaction of sugars and amino acids in determining the origin of chirality, Special Feature: The 41st Symposium, Viva Origino, 44, 4.
- Tamura K, Schimmel P (2004), Chiral-selective aminoacylation of an RNA minimax. Science 305.
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K,. Sugino A (1968) Mechanism of DNA chain growth. I: possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci USA, 59: 598-605.
- Karasawa S (1974) Origin of Piezoelectricity in an α-Quartz, Japanese. Journal of Applied Physics 13: 799-803.
- Karasawa S (2022) Earliest BIF and life produced via submarine volcanism in carbonated seawater. Geology, Earth & Marine Sciences 4: 1-5.
- Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM (1987) Molecular Biology of the gene [4th ED] Chapt.9, p.241, The Benjamine/Cumming Publishing Company, Inc., USA.
- McKee T, McKee JR (2016) Biochemistry the Molecular Basis of Life, 6th, Sec.18.1, Oxford Univ. Press, UK.
Adhered Sugars became a Component of Nucleic Acids
The Structure of Phospholipid Membrane
The current cell membrane has a hydrophilic “head” containing a phosphate group and two hydrophobic “tails” derived from fatty acids, joined by a glycerol molecule as shown in Figure 3.
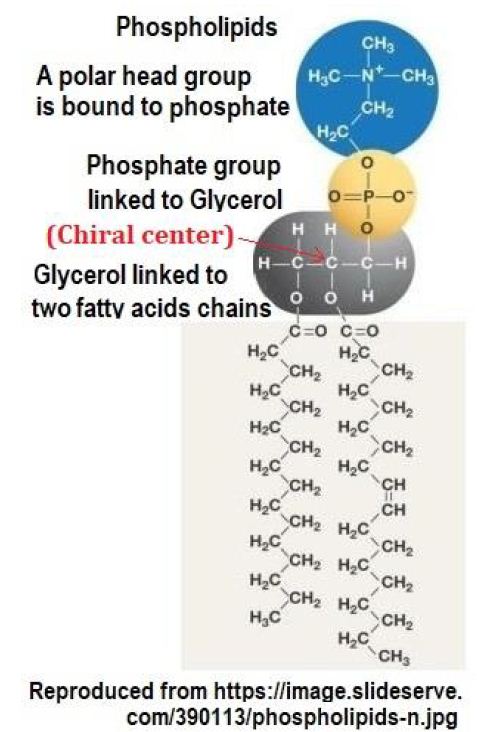
Figure 3: The structure of phospholipid membrane. Phospholipid arrangement in cell membranes contains a chiral center at the C2 position of glyceryl moiety.
A phospholipid contains a chiral center at the C2 position of glyceryl moiety. The twisted phospholipids will interlock, and the interlocked molecules induce systematic motions of the same kind chiral molecules due to systematic thermal vibrations of atoms in the molecule.
Screw Movement in a Phospholipid Bilayer
In case that the bilayer of which one of the layers is rotated by 180°, the progress of helix is changed from output side to input side at the center of the bilayer as illustrated in Figure 4. Despite this bilayer having two chirality centers, the bilayer provides a one directional screw movement by the one directional rotation.
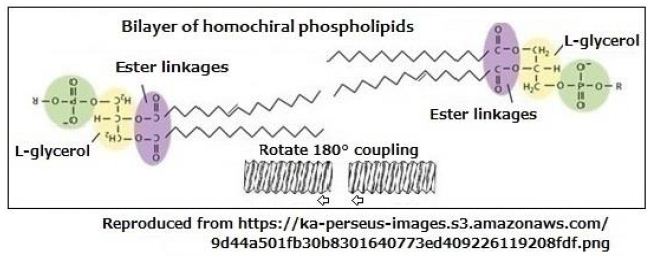
Figure 4: The bilayer of which one of the layers is rotated by 180° where a progress of screw movement continues from output side to input side at the center of the bilayer.
Here, there are two kinds of helical structures concerned with rotational direction of screw movements. When L-type of amino acids inserted in the phospholipid’s bilayer, L-type of bilayer will be formed, and D-type of sugar molecules proceeds in emitting direction along the induced L-type bilayer. The chirality of molecule that is synthesized by insertion and the molecule that is emitted by synthesize are different.
How DNA Structure was Formed
How mRNA and tRNA had Formed
The evolution of repetitive productions of useful proteins has added the process of regenerating proteins using RNA precursors. The decompose shortly RNA of a tool for replication is favorable for life. So, DNA suitable for memory needs to be synthesized. The tool for replication of protein was replicated from DNA. The evolved process of replication of protein is carried out by mRNA and tRNA with support of plural of complicated enzymes, where amino acid is carried by tRNA, and those amino acids are arranged to a protein by the information of mRNA. Each step of matching between codon and anticodon was checked by as if a contact point of gear mechanism. The lagging strand that adhered with amino acid makes link to corresponding portion of mRNA by using genetic codes. Since the chirality of amino acid adhered to leading strand is L-type, the new leading strand fits with L-typed surrounding molecules and it will be a long strand. On the other hand, the chirality of new lagging strand is D-type, and it is easily leaved from surrounding molecules, and it will become short strands.
Structure of Double Helix of DNA
DNA is made of chemical building blocks of nucleotides. Each block is made of three parts: a phosphate group, a sugar group and nitrogen bases. The phosphate and sugar groups are linked into chains alternatingly, and a pair of strands is coupled by nitrogen bases [13,14]. There are rotational thermal vibrations of atoms in the helical structure of molecules. Sugar molecules are possible to adhere on the surface of the membrane, and those will be linked by phosphate. Strands of DNA will be formed along the rotating helical structures of the surface as shown in Figure 2. The synthesis of strand is facilitated by nucleophilic action of 3′ hydroxy group (-OH) of the terminal residue to phosphate group (PO4). Then, carbon 3′ will be adhered to carbon 5′ by phosphate-mediated ester bonds. The growing of leading strand proceeds continuously repeatedly from the 5′ to 3′ end. L-type of helical structure of strand in a cell are induced in the vicinity of the protein.
How Genetic Codes had Formed
Two of strands in a DNA are linked by bases. The strands of DNA are coupled by nitrogen bases by hydrogen bond at each synthesis steps, naturally generated nitrogen base pairs will be mixture of various combinations. As the result of natural selection, the linkage of strands favorable for survival was remained. Bases of current DNA are adenine (A), cytosine (C), guanine (G) and thymine (T). There are two kinds of pairing bases those have complementary stable pairs. One of the pair is A (adenine) and T (thymine), and the other is C (cytosine) and G (guanine). The one amino acid is assigned by three set of bases among 4 set of bases. Since tRNA must link with anticodon and amino acid, structure of tRNA becomes complicated. “Aminoacyl tRNA synthetase” binds the amino acid to specific tRNA that identifies their corresponding codons and “ribosome” ensures that tRNA correctly identifies mRNA. Although the mechanism of tRNA is complicated, the resultant role of tRNA is simple. An illustration on the stepping proceedings of activated portions of leading strand and lagging strand are shown in Figure 5.
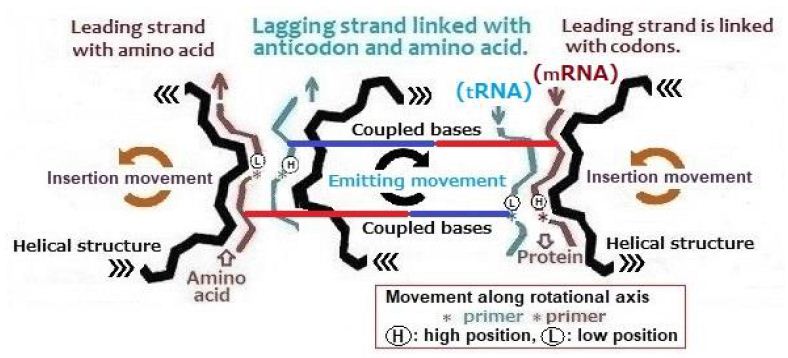
Figure 5: Proceeding of activated paired bases in a double helix DNA
Enzymes of “Helicase” and “Polymerase” for Replication of DNA
Current replication of DNA is processed by using existing DNA. Depending on the twisting direction, the joint of nitrogen base of DNA can be separated, or that can be brought closer. The motion of twist cannot achieve without any change of the surrounding molecules. Since the system of molecules to drive the twist motion includes amino acids and the amino acids make protein, enzyme for the function of twist is reproduced by the protein. The current DNA system makes use of helicase to separate two strands of a DNA to join and polymerase to join two stands. Helicase continues to move toward the unelicited double-stranded region, and leaved two single-stranded DNA templates that induce the formation of double-stranded daughter DNA duplexes. Since the separated strand can only be synthesized in the direction of extending the 3′ end, only the leading strand can continuously replicate as helicase moves.
The processes of current replication of DNA are as follows.
The representation on replication of DNA is shown in Figure 6. The proceeding of active portion in lagging strand rotates in the opposite for that of leading strand, and the synthesizing DNA waits for some parts of strand to be exposed and continues until it meets the 5′ end of the previously synthesized lagging strand. A short DNA fragment formed by the lagging strand is called “Okazaki fragment” [10]. The Okazaki fragment is connected to the next Okazaki fragment shortly after synthesis to form a continuous new DNA. The different chirality of strands is necessary because reproduced two DNA’s must be divided into two equal parts in the beginning processing of a cell division.
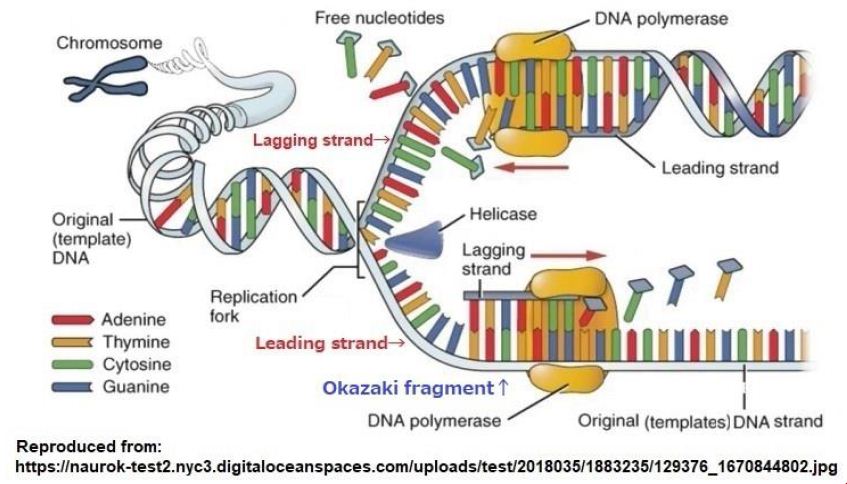
Figure 6: Replication of DNA by using original DNA
Conclusion
This paper described a hypothesis about how DNA may have evolved in the water environment of the Earth. The interactions among molecules in DNA are able to get hints from stereo structure of DNA. There are leading strand and lagging strand in a DNA. The proceeding of lagging strand is rotationally opposite direction to that of leading strand. The strands with different chirality in a DNA are coupled by nitrogen bases through hydrogen bond at each synthesis steps. When DNA is first created, every element that makes up the mechanism must be incorporated at the same time. It is known that L-type chirality of amino acid had been selected, and L-type of helical movements of molecules are able to assist to synthesize L-typed helical filamentous molecules. It can be considered that D-type of helical movements will be induced through interactions among L-type of helix via gear mechanism. Based on such estimations, it is possible to explain the generation of D-type of sugar of DNA on the surface of cell membrane. The first formation must be achieved tremendous number of try and errors. Current process of replicating DNA uses existing DNA. After separation of paired strands of DNA, the proceeding of leading strand is rotationally opposite to that of lagging strand, it makes possible to check of key-lock relationship on codon and anticodon. Replication of a protein involves multiple reactions. As further prospect, an enzyme is made by protein. However, our understanding is causal law, we cannot express precise progress of concurrent different reactions by using logical expression. Overlapping multiple causal laws can be expressed by using a pattern. Although there exists unknown mechanisms, to reveal various aspects of surroundings contributes to improve the understanding of the complicated mechanism of protein replication. It can be concluded that the chirality of biomolecule is able to take a role of bridge between origin of life and the molecular biology.
Acknowledgement
The author expresses his sincere thanks to Professor Koji Tamura from Tokyo University of Science for helpful discussions and comments on the manuscript.
References