Abstract
SARS-COV-2 is the etiologic agent of COVID-19. There is currently no effective remedy for SARS-COV-2 infections or COVID-19. Dimerization of SARS-COV-2 encoded Nucleocapsid protein (NCp) is a prerequisite step for it to act as an essential co-factor for the replication, transcription and packaging of SARS-COV-2 genome. Molecules that prevent the dimerization of NCp are potential prophylactics and therapeutics for the control of SARS-COV-2 infections and virulence. Here, through interrogation of chemical ligand data banks and thermodynamic calculations, we show that Chlorophyll A, Halothane and Tetraethylene glycol monooctyl ether (TGME) are inhibitors of the dimerization of NCp. Chlorophyll A is the most potent inhibitor of NCp dimerization with dissociation constant (KD) of ~28 pM. Chlorophyll A binding caused the dissociation constant (KD) for NCp-NCp interaction to increase from ~7.2 pM to ~1000000 pM. Chlorophyll A also bound to NCp mutated at phosphorylation sites S186, S197 and S202 (S186F, S197L and S202N) and phosphorylation recognition sites RNpSTP, (S197L) and RGTpSP (RG203/204KR and RG203/204KT) with dissociation constants of ~12 pM, ~6.1 pM, ~27.8. pM, ~27.8 pM and ~2.2 pM respectively. These results show that Chlorophyll A, a chemical ligand that is present in high abundance with good absorption properties and near-zero toxicity is a potential very potent prophylactic and therapeutic that acts via disruption of NCp dimerization.
Introduction
SARS-COV-2 is the etiologic agent of COVID-19 [1-4], a highly debilitating disease of the respiratory system that has so far killed in excess of 1 million individuals (still counting) worldwide [5,6]. There is currently no known cure for COVID-19. Although, there are press releases of candidate vaccines that are more than 90% effective in preventing symptoms of COVID-19, there are no vaccines for the control of SARS-COV-2 infections [6-11]. In order to prevent SARS-COV-infections and virulence of 2, the replication cycle of SARS-COV-2 must be inhibited. Among the SARS-COV-2 encoded proteins [1-4], the Nucleocapsid protein (NCp) has an essential role in the initiation and control of the replication, transcription and packaging of the SARS-COV-2 genome [12-23]. Dimerization and oligomerization of SARS-COV-2 Nucleocapsid protein (NCp) is a pre-requisite for NCp to act as a co-factor for the initiation and control of replication, transcription and packaging of of the SARS-COV-2 genome [16-23]. Targeting NCp dimerization is an attractive avenue for a drug discovery program that aims to block the replication, transcription and packaging of the SARS-COV-2 genome. Molecules that act to prevent dimerization of NCp are prime therapeutic candidates Because of its critical function in the replication, transcription and packaging of the SARS-COV-2 genome, it is not surprising that NCp has been considered a prime target for drug discovery programs [24-26]. NCp has also been proposed as a vaccine target [27-32]. NCp is phosphorylated at multisite sites which control its function [20,33-38]. However, NCp has been shown to undergo mutations at several of its phosphorylations sites [33]. While phosphorylation of NCp at key sites within a phosphorylation rich domain has been proposed to act as a cellular response mechanism for phosphorylation dependent sequestration of NCp by Protein 14-3-3, mutations in the phosphorylation sites appear to be viral mechanism for avoiding sequestration of NCp by Protein 14-3-3 [30,38]. Mutations of phospho-sites including serine 186 (S186F), serine 197 (S197L), serine 2020 (S202N) and phosphorylation recognition sites RG 203/204 (RG203/204KR and RG203/204KR) have been shown to occur in strains/sub-strains of SARS-COV-2 that were isolated in various populations around the world [30,38]. Any molecules that bind to NCp to disrupt its dimerization must not only bind to the wild-type NCp but also to the mutated NCps. In this work, it is shown that a chemical ligand, Chlorophyll fulfills this requirement and that Chlorophyll A bound NCp and mutated NCps at picomolar concentration and causing major increases in the dissociation constants (KDs) for the wild-type and mutated NCp-NCp dimer interactions.
Methods
The structure of dephospho-SARS-COV-2 Nucleocapsid protein (NCp) was rendered de novo using the Quark Program pursuant to Zu and Zhang [39,40]. The identification of a chemical ligand that binds to NCp was performed by interrogating Chemical Ligand Data Banks using the Coach Program pursuant to Yang et al. [41,42]. Each chemical ligand that was identified was then analyzed for its ability to bind NCp. Renaming of chains in NCp-chemical ligand and NCp-chemical ligand-NCp complexes were performed by using the Rename Chain PDB File Program pursuant to Rath, E. [43]. Mutations of the phosphorylation sites and phosphorylation recognition sites of NCp was performed using the Build Model Program of FoldX pursuant to Guerois et al. and Schymkowitz et al. [44,45]. The bindings of identified Chemical ligands-NCps complexes to wild-type NCp and mutated NCps were analyzed by Docking Experiments using Z Dock Program pursuant to Pierce et al. [46]. Monomeric and dimeric wild-type and mutated NCps rendered in this work were analyzed and visualized by the CCP4 Molecular Graphics Program Version 2.10.11 as described by Mc Nicolas et al [47] and the ZMM Molecular Modeling Program as described by Garden and Zhorov [48]. The dissociation constants (KDs) for the binding of chemical ligands to NCp monomer and dimers was determined by first obtaining the Gibbs Free Energy (ΔGGFG) using the Prodigy-Ligand Program pursuant to Vangone et al. and Kurkcuoglu et al. [49,50] and then calculating KDs from the equation KD = e(-ΔG/RT). The dissociation constant (KD) for the NCp-NCp interactions in NCp dimers were determined by using the Prodigy-Protein-Protein Program pursuant to Vangone and Bonvin, and Xue et al. [51,52].
Results
The structure of dephospho NCp (amino acids 123-310) which encompasses the phosphorylation rich domain of NCp (amino acids 185-209) was rendered de novo using the Quark Program pursuant to Xu and Zhang [39,40]. Figure 1 shows the structure of the rendered monomeric structure of NCp. The structure of the dimeric NCp was determined by Docking Experiments and is summarized in Figure 2. Monomeric NCp was used to interrogate Chemical Ligand Data Banks using the COACH program pursuant to Yang et al. [41,42]. A number of Chemical Ligands that bound to NCp, including Halothane, Tetraethylene glycol monooctyl ether (TGME) and Chlorophyll A were identified (Figure 3). The 3 chemical ligands bound monomeric NCp with dissociation constants (KDs) of ~66 µM, ~78 nM and ~28 pM respectively and dimeric NCps with dissociation constants (KDs) of ~66 µM, ~24 µM and ~12 pM respectively (Figure 4). The binding of Halothane to dimeric NCps was accompanied by an increase of the dissociation constant (KD) for the NCp-NCp interaction from 7.2 pM to 150 pM. The binding of TGME caused the dissociation constant (KD) for the NCp-NCp interaction to increase from 7.2 pM to 4000 pM while the binding of Chlorophyll A resulted in an increase of the dissociation constant (KD) for the NCp-NCp interaction from 7.2 pM to 1000000 pM. The binding of Chlorophyll A to NCp was further characterized because of its very high affinity binding to NCp (~28 pm for monomeric NCp and ~12 pM for dimeric NCp) and extremely profound effect on the structure, conformation (Figures 3-6) and dissociation constant (KD) for the NCp-NCp interaction (from 7.2 pM to 1000000 pM). Chlorophyll A formed contact points with amino acid residues, Tyrosine 50, Glutamic acid 52, Glycine 53, Arginine 55, Glycine 56, Glutamine 59, Serine 61, Serine 62, Arginine 63, Serine 66, Leucine 100, Leucine 101, Aspartic acid 103, Arginine 104, Lysine 135, Threonine 141, Alanine 142, Tyrosine 146, histidine 176, Glutamine 181, Isoleucine 182, Alanine 183, Alanine 186, Threonine 174, Alanine 178, Glutamine 184, Phenylalanine 185 and Proline 187 of NCp (Figures 5A and 6A) and also with the phosphorylation rich domain of NCp (Figure 5B and 6B). NCp has been shown to be mutated at several phosphorylation sites and phosphorylation recognition sites within the phosphorylation domain of NCp, including serine 186 (S186F), serine 197 (S197L), serine 202 (S202N), Arginine 203 and Glycine 204 (RG203/204KR and RG203/204) [33,38]. It was previously proposed that cells infected with SARS-COV-2 possess a cellular response mechanism for the binding and sequestration of NCp by Protein 14-3-3 involving multi sites phosphorylation by a variety of cellular protein kinases, and in counterpart, SARS-COV-2 has evolved to evade the cellular response mechanism through mutations of at least 3 phosphorylation sites, serines 186, 197 and 202 and 2 phosphorylation recognition sites (RNpSTP and RGTpSP), serine 197, arginine 203 and glycine 204 [33,38]. It was therefore necessary to determine whether Chlorophyll A can bind all the NCp mutants with high affinities. The dissociation constants (KD) for the binding of Chlorophyll A to wild type and mutated monomeric and dimeric NCps are summarized in Tables 1 and 2. These results show that Chlorophyll A bound monomeric and dimeric wild type and mutant NCps with very similar high affinities.
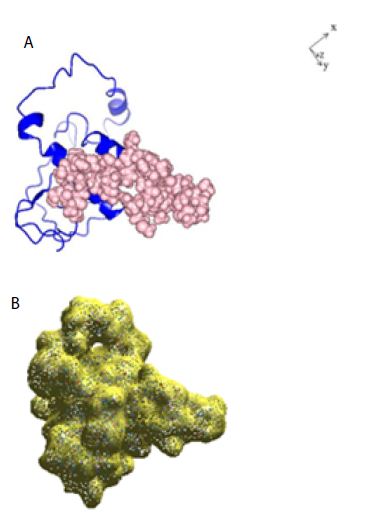
Figure 1: A: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein monomer (NCp monomer), rendered as described in Method Section. The amino acid sequence of the phosphorylation rich domain is depicted as spheres in pink. B: Realistic rendering of SARS-COV-2 Nucleocapsid protein (NCp) monomer.
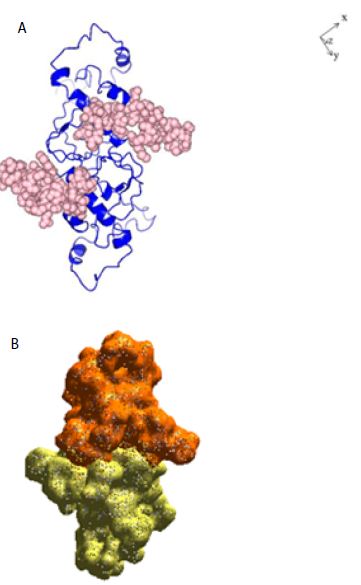
Figure 2: A: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein dimer (NCp dimer), rendered as described in Method Section. The amino acid sequence of the phosphorylation rich domain is depicted as spheres in pink. B: Realistic rendering of SARS-COV-2 Nucleocapsid protein (NCp) dimer.
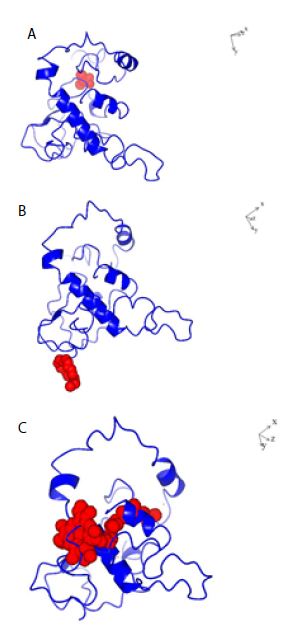
Figure 3: Structures of SARS-COV-2 Nucleocapsid protein monomer (NCp monomer) with bound chemical ligands. A; Halothane (Red); B: Tetraethylene glycol monooctyl ether (Red); C: Chlorophyll (Red), The dissociation constants (KDs) for the binding of NCp monomer-chemical ligands were 66 µM, 78 nM and 28 pM for Halothane, Tetraethylene glycol monooctyl ether and Chlorophyll A respectively.
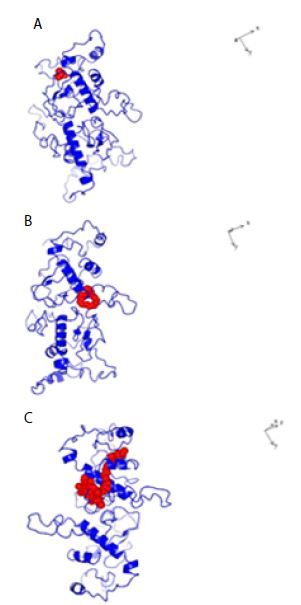
Figure 4: Structures of Dimeric SARS-COV-2 Nucleocapsid protein monomer (NCp monomer) with bound chemical ligands. A; Halothane (Red); B: Tetraethylene glycol monooctyl ether (Red); C: Chlorophyll (Red), The dissociation constants (KDs) for the binding of NCp dimer and chemical ligands were 66 µM, 24 µM and 28 pM for Halothane, Tetraethylene glycol monooctyl ether and Chlorophyll A respectively.
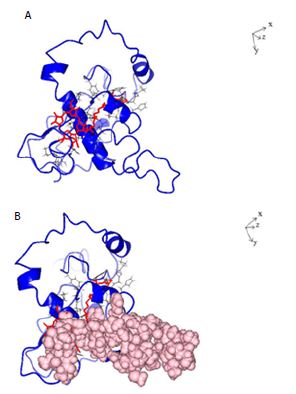
Figure 5: A: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein monomer (NCp monomer) bound by Chlorophyll A (Red), rendered as described in Method Section. The contact points in NCp are shown in Black. B: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein monomer (NCp monomer) bound by Chlorophyll A (Red), The contact points in NCp are shown in Black. The phosphorylation rich domain of NCp is depicted as spheres in pink.
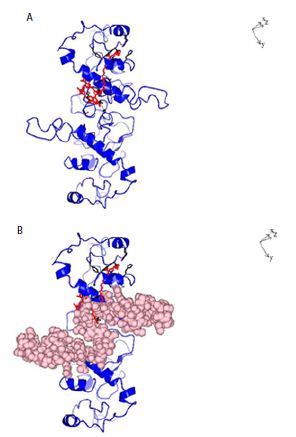
Figure 6: A: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein dimer (NCp dimer) bound by Chlorophyll A (Red), rendered as described in Method Section. The contact points in NCp are shown in Black. B: Ribbon structure (blue) of SARS-COV-2 Nucleocapsid protein dimer (NCp dimer) bound by Chlorophyll A (Red), The contact points in NCp are shown in Black. The phosphorylation rich domain of NCp is depicted as spheres in pink.
Table 1: Dissociation constants (KDs) for the binding of Chlorophyll A, Halothane and Tetraethylene glycol monooctyl ether (TGME) to NCp monomer and NCp dimer, and dissociation constants (KDs) for NCp-NCp interactions in the presence of Chlorophyll A, Halothane and Tetraethylene glycol monooctyl ether (TGME).
|
Dissociation constant (KD) for Ligands binding (pM) |
Dissociation constant (KD) for NCp-NCpinteractions in NCp dimer (pM) |
|
|
-NCp |
~00.0 |
– |
|
NCp-NCp Complex |
– |
~7.2 |
|
-NCp-Chlorophyll A complex |
~28.0 |
– |
|
-NCp-NCp-Chlorophyll A Complex |
~12 |
~1000000 |
|
-NCp-Halothane Complex |
~66000000 |
– |
|
-NCp-NCp-Halothane Complex |
~66000000 |
~150 |
|
-NCp-TGME Complex |
~2400000 |
– |
|
-NCp-NCp-TMGE Complex |
~78000 |
~4000 |
Table 2: Dissociation constants (KD) for the binding of Chlorophyll A to NCp monomer, NCp monomer mutants, NCp dimer and NCp dimer mutants.
|
|
Dissociation constants (KDs) for Chlorophyll A binding (pM) |
|
Wild Type-NCp-Chlorophyll A complex |
~28.0 |
|
S186F mutant-NCp-Chlorophyll A complex |
~6.0 |
|
S197L mutant-Chlorophyll A Complex |
~6.1 |
|
S202N mutant-NCp-Chlorophyll A complex |
~27.8 |
|
RG203/204KR mutant-NCp-Chlorophyll A complex |
~27.8 |
|
RG203/204KT mutant-NCP Chlorophyll A complex |
~6.1 |
|
Wild Type-NCp-NCp complex |
– |
| Wild Type-NCp-Chlorophyll A-Wild Type NCp complex |
~12.0 |
|
S186F mutant-NCp-Chlorophyll A-S186F mutant-NCp complex |
~1.0 |
| S197L mutant-NCp-Chlorophyll A-S197L mutant-NCp Complex |
~6.1 |
|
S202N mutant-NCp-Chlorophyll A-S202N mutant-NCp complex |
~27.8 |
| RG203/204KR mutant-NCp-Chlorophyll A-RG203/204KR mutant-NCp complex |
~27.8 |
|
RG203/204KT mutant-NCp Chlorophyll A-RG203/204KT mutant-NCp complex |
~2.2 |
Discussion
There is currently no effective means to control SARS-COV-2 infection and virulence. There is also no cure for COVID-19, the disease(s) caused by SARS-COV-2. Although, there are press releases describing 3 candidate vaccines to be over 90% effective, it is clear that they do not prevent SARS-COV-2 infections [7-11]. Full disclosure of the clinical trials of the effectiveness of the 3 candidate vaccines is required before it can be determined scientifically with some degree of certainty that they are indeed effective as prophylactics. The effective control of SARS-COV-2 infection and virulence can be achieved through a disruption of the replication, transcription and packaging of the SARS-COV-2 genome. NCp is an essential co-factor in the replication, transcription and packaging of the SARS-COV-2 genome [12-23]. Inhibiting the function of NCp is therefore a very attractive way to prevent the viability, transmission, infection and virulence of SARS-COV-2 [24-26]. Dimerization and oligomerization are prerequisite steps that enable NCp to act as an essential co-factor in the replication, transcription and packaging of the SARS-COV-2 genome [12-23]. In the present work, a number of chemical ligands, including Halothane, Tetraethylene glycol monooctyl ether (TGME) and Chlorophyll A have been identified and shown to inhibit the dimerization of NCp. All 3 molecules bind to NCp with high affinities, with dissociation constants (KDs) of 66 µM, 78 nM and 28 pM respectively, and disrupts the interaction between NCps by causing the dissociation constant (Kd) of the NCp-NCp interaction to significantly increase from ~7.2 pM to 150 pM, 4000 pM and 1000000 µM respectively. The effects of the chemical ligands on the dimerization of NCp are therefore profound. It was previously shown that NCp becomes mutated at key phosphorylation sites, including serine 186 (S186F), serine 197 (S197L) and serine 202 (S202N) and phosphorylation recognition sites, serine 197 (S197L), arginine 203 and glycine 204 (RG203/204KR and RG203/204KT) within the motifs, RNpSTP and RGTpSP that are located in the linker region of NCp [33,38]. It has been proposed that cells infected with SARS-COV-2 possess a cellular response mechanism for the binding and sequestration of NCp by Protein 14-3-3 involving multi sites phosphorylation by a variety of cellular protein kinases and in counterpart, SARS-COV-2 has evolved to evade the cellular response mechanism through mutations of at least 3 phosphorylation sites, serines 186, 197 and 202 and three phosphorylation recognition sites (RNpSTP and RGTpSP), serine 197, arginine 203 and glycine 204 [33,38]. In the present work, it is shown that Chlorophyll A bound to all the relevant NCp mutants with high affinities. It is submitted that Chlorophyll A is a very potent inhibitor of the dimerization of SARS-COV-2 encoded Nucleocapsid protein (NCp). Because of its relative abundance, good absorption properties and near zero toxicity, the development of Chlorophyll A as a prophylactic and therapeutic for the control of SARS-COV-2 infections and virulence is warranted. In contrast to candidate vaccines that can only prevent symptoms of COVID-19 [7-11], Chlorophyll A will not only act as a prophylactic and therapeutic to cure COVID-19, it will also prevent SARS-COV-2 infection and viability because it acts via inhibition of the dimerization of NCp, an essential and prerequisite step in the initiation and control of the replication, transcription and packaging of the SARS-COV-2 genome.
Acknowledgement
This work was supported by the Nacbraht Biomedical Research Institute Fund.
Author Contribution
H.Y. Lim Tung came up with the concept and the questions, performed the experiments with Pierre Limtung, analyzed the results with Pierre Limtung and wrote the paper with Pierre Limtung.
Pierre Limtung performed the experiments with H.Y. Lim Tung, analyzed the results with H.Y. Lim Tung and wrote the paper with Pierre Limtung.
Conflict of Interest
The authors have no conflict of interest to declare.
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727-733. [crossref]
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395: 565-574. [crossref]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273. [crossref]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269. [crossref]
- Tung HYL (2020) SARS-COV-2 infection and virulence: classification of seven groups of countries. IP 10000 (infectivity) and DP 10000 (virulence) are influenced by temperature, humidity and far infrared irradiation. New Biomed Sci 1: 71-85.
- Tung HYL (2020) Faulty response strategy and abject failure to control SARS-COV-2 infections and virulence. J Invest Cri Pub Sci
- Pfizer (2020) Pfizer and BioNTech announce vaccine candidate against COVID-19 achieved success in first interim analysis from phase 3 study. Pfizer.com, Issue of November 9, 2020.
- Calloway E (2020) What Pfizer’s landmark COVID vaccine results mean for the pandemic. Nature, Issue of November 9, 2020.
- Cohen J (2020) Champagne and question greet first data showing that a COVID-19 vaccine works. Science, Issue of November 9, 2020.
- Cohen J (2020) ‘Just beautiful’: Another COVID-19 vaccine from newcomer Moderna, succeeds in large scale trial. Science Issue of November 16, 2020.
- Calloway E (2020) COVID Vaccine excitement builds as Moderna reports third positive result. Preliminary data show that the immunization is 94% effective and seems to prevent severe infections. Nature, Issue of November 16, 2020.
- Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido, et al. (2007) The intracellular sites of early replication and budding of SARS coronavirus. Virology 361: 304-315. [crossref]
- Verheije MH, Hagemeijer MC, Ulasli M, Reggiori F, Rottier PJ, et al. (2010) The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J Virol 84: 11575-11579. [crossref]
- Cong Y, Ulasli M, Schepers H, Mauthe M, V’kovski P, et al. (2020) Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J Virol DOI: 10.1128/JVI.01925-19.
- Tylor S, Andonove A, Cutts T, Cao J, Grudesky E, et al. (2009) The SR-rich motif in SARS-CoV nucleocapsid protein is important for virus replication. Can J Microbiol 55: 254-260. [crossref]
- Chen CY, Chang CK, Chang YW, Sue SC, Bai, et al. (2007) Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol 368: 1075-1086. [crossref]
- Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, et al. (2005) Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol 79: 13848-13855. [crossref]
- Yu IM, Gustafson CLT, Diao J, Burgner II, et al. (2005) Recombinant Severe Acute Respiratory Syndrome (SARS) Coronavirus Nucleocapsid Protein Forms a Dimer through Its C-terminal Domain. J Biol Chem 280: 23280-23286. [crossref]
- He R, Dobie F, Ballantine M, Leeson A, Li, Y, et al. (2004) Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun 316: 476-483. [crossref]
- Peng TY, Lee, K.-R,Tarn WY (2008) Phosphorylation of the arginine/serine dipeptide‐rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J 275: 4152-4163. [crossref]
- Surjit M, Liu B, Kumar P, Chow VT, Lal S K (2004) The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem Biophys Res Commun 317: 1020-1036. [crossref]
- Luo H, Ye F, Sun T, Yue L, Peng S, et al. (2004) In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys Chem 112:15-25. [crossref]
- Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, et al. (2005) The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett 579: 5663-5668. [crossref]
- Kang S, Yang M, Hong Z, Zhang L, Huang Z, et al. (2020) Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharma Sinica 10: 1228-1238. [crossref]
- Mukherjee D, UPASANA R. (2020) SARS-CoV-2 Nucleocapsid Assembly Inhibitors: Repurposing Antiviral and Antimicrobial Drugs Targeting Nucleocapsid-RNA Interaction. chemRxiv, DOI: 10.26434/chemrxiv.12587336.v2.
- Yadav R, Imran M, Dhamija P, Kapil Suchal K, Handu, S. (2020) Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J Biomol Struc Dynamics, DOI: 1080/07391102.2020.1778536. [crossref]
- Gao W, Tamin A, Soloff A, D’Aiuto L, Nwanegbo E, et al. (2003) Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet 362:1895-1896. [crossref]
- Liu SJ, Leng CH, Lien SP, Chi HY, Huang CY, et al. (2006) Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24: 3100-3108.
- Lin Y, Shen X, Yang RF, Li YX, Ji YY, et al. (2003) Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res 13: 141-145. [crossref]
- Peng H, Yang LT, Wang LY, Li J, Huang J, et al. (2006) Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients Virology 351: 466-475. [crossref]
- Kalita P, Padhi AK, Zhang K, Tripathi, T. (2020) Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microbial Pathogenesis, DOI: 10.1016/j.micpath.2020.104236
- Dutta NK, Mazumdar K, Gordy JT (2020) The Nucleocapsid Protein of SARS–CoV-2: a Target for Vaccine Development. J Virol 94. [crossref]
- Tung HYL, Limtung Pierre (2020) Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein and structure model of sequestration by protein 14-3-3. Biochem Biophys Res Commun 532: 134-148. [crossref]
- Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, et al. (2005) The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol 79:11476-11486. [crossref]
- CH Wu, SH Yeh, YG Tsay, et al. (2009) Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem 284: 5229-52239. [crossref]
- Wu CH, Chen PJ, Yeh SH (2014) Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables Corona virus transition from discontinuous transcription. Cell Host and Microbe 16: 462-472. [crossref]
- Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, et al. (2020) Phosphorylation modulates liquid-liquid phase separation of the SARS-CoV-2 N protein. bioRxiv, DOI: 10.1101/2020.06.28.176248..
- Limtung P, Tung HYL (2020) Structure Model Analysis of the Effects of Mutations in the Phosphorylation Sites of SARS-CoV-2 Encoded Nucleocapsid Protein. Sci J Biol, DOI: 37871/sjb.id19.
- Xu D, Zhang Y (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80: 1715-1735. [crossref]
- Xu D, Zhang Y (2013) Toward optimal fragment generations for ab initio protein structure assembly. Proteins 81: 229-239. [crossref]
- Yang J, Roy A, Zhang Y (2013) BioLiP: a semi-manually curated data base for biologically relevant ligand protein interactions. Nucleic Acids Res 41:1096-1103. [crossref]
- Yang J, Roy A, Zhang Y (2013) Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 20: 2588-2595. [crossref]
- Rath E (2010) https://canoz.com/sdh/renamepdbchain.pl.
- Guerois R, Nielson JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320: 369-387. [crossref]
- Joost Schymkowitz J, Jesper Borg J, Francois Stricher F, Robby Nys R, Frederic Rousseau F, et al. (2005) The FoldX web server: an online force field. Nucleic Acids Res 33: W382-W388. [crossref]
- Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, et al. (2014) ZDOCK Server: Interactive Docking Prediction of Protein-Protein Complexes and Symmetric Multimers. Bioinformatics 30: 1771-1773. [crossref]
- McNicholas S, Potterson E, Wilson KS, Noble MEM (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Cryst D67: 386-394. [crossref]
- Garden DP, Zhorov BS (2010) Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J Comp Aided Mol Des 25: 91-105. [crossref]
- Vangone A, Schaarschmidt J, Koukos P, Geng C, Citro N, et al. (2018) Large-scale prediction of binding affinity in protein–small ligand complexes: the PRODIGY-LIG web server. Bioinformatics 35: 1585-1587.
- Kurkcuoglu Z, Koukos PI, Citro N, Trellet ME, Rodrigues JPGLM, et al. (2018) Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. J Comp Aid Mol Des 32:175-185.
- Vangone A, Bonvin AMJJ (2015) Contact based prediction of binding affinity in protein-protein complexes. eLife [crossref]
- Xue L, Rodrigues J, Kastritis P, Bonvin AMJJ, Vangone A (2016) PRODIGY: a webserver for predicting the binding affinity in protein-protein complexes. Bioinformatics 32: 3676-3678. [crossref]