Abstract
A highly selective Salmonella and Shiga Toxin-producing E. coli (STEC) enrichment medium broth (SSS; commercially known as PDX-STEC), in comparative studies of low level E. coli O157:H7 inoculated ground beef and spinach, showed a 50- to 100- fold increase in STEC recoveries of the pathogen from ground beef compared to spinach enrichments. These observations suggested that either a soluble component of spinach inhibited the growth of the E. coli O157:H7 or a soluble component of ground beef stimulated the growth of the pathogen. The growth stimulating effect was linked to a soluble component of ground beef by comparing the growth of STEC and Salmonella in SSS conditioned with ground beef by passive extraction to their growth in control SSS containing traditional powdered beef extract media supplement. Then attempts were made to isolate and identify the responsible compound(s). A 20 to 60% ammonium sulfate fraction of ground beef extract maintained the growth stimulation of STEC and Salmonella. Further purification using affinity chromatography and preparative polyacrylamide gel electrophoresis identified three specific protein bands (52 kD, 35 kD and 20 kD) associated with the growth stimulating activity. Mass spectral analysis of the trypsin-digested peptides of these proteins provided a putative identification of the proteins as the glycolytic protein, phosphoglucomutase (E.C.5.4.2.2). Finally, commercially prepared rabbit muscle Phosphoglucomutase (PGM) was shown to have the same growth stimulating activity thereby confirming the identity of the active protein. The possible mechanisms of growth stimulation by PGM may be through increasing bacterial fitness and environmental adaptability. Inclusion of PGM in food safety test protocols can enhance detection and isolation of contaminating STEC and Salmonella.
Highlights/Significance
- A putative growth stimulating factor was linked to 20 kD, 35 kD and 52 kD proteins.
- Mass spectral analysis provided a provisional identification of the protein as Phosphoglucomutase (PGM) which was then confirmed using a commercially available PGM.
- This protein can be used as an enrichment media supplement to improve the detection and recovery of STEC and Salmonella in foods.
Keywords
Food safety, Shiga toxin-producing E. coli, Salmonella, Selective enrichment
Introduction
Food borne illness linked to Shiga Toxin-producing Escherichia coli (STEc) and Salmonella enterica is on-going problem in the United States. United States Department of Agriculture (USDA) Food Safety and Inspection Service (FSIS) recalls involving these pathogens were reported in 2019 and 2020 in various food stuffs [1-3]. The US Food and Drug Administration (FDA) initiated recalls during the same period involved cantaloupe [4], cinnamon apple chips [5], peaches [6] and flour [7]. The frequency and breadth of food stuffs contaminated with STEC and Salmonella demonstrate that there are challenges in the available testing methodologies to properly assess the food safety systems producing these food products. The challenges in food testing methodologies may permit contaminated food to enter the national food supply.
E coli is usually a harmless bacterium living in the gastrointestinal tract of humans and other mammals. There are different serotypes of E. coli that have acquired bacterial Shiga toxin genes, originally arising in Shigella. The most prominent STEC associated with severe human disease in the US is E. coli O157:H7. This serotype is associated with cattle, its natural reservoir and can contaminate beef products during harvest and processing. Six other serogroups of STEC (O26, O45, O103, O111, O121 and O145) are responsible for about three-quarters of non-O157 STEC illness in the US [8]. These multiple serotypes and serogroups of pathogenic E. coli can be indistinguishable from the harmless E. coli of the gastrointestinal tract and thus pose a challenge for detection and isolation. Likewise, the CDC has identified Salmonella serotypes Enteritids, Newport, Typhimurium, and Javiana as the most common serotypes causing reportable Salmonellosis [9]. Although these are the most frequently reported Salmonella serotypes overall, their attribution to various food stuffs varies [10], with serotypes Typhimurium and Newport most commonly attributed to beef, while Enteritidis is attributed to chicken and eggs, and less common serotypes like Heidelberg attributed to turkey products. Even uncommon Salmonella serotypes such as Tennessee have been observed to emerge as significant outbreak strains, as occurred in 2006 associated with peanut butter [9].
The technical challenges of distinguishing these gram-negative pathogens from harmless gastrointestinal coliforms has been rendered substantially less difficult with the advent of PCR screening methods that target specific portions of the E. coli O157:H7 genome or common virulence factors such as the Shiga toxin gene (stx) present in STEC [11]. Similar molecular methods targeting virulence markers such as the invasion gene (invA) of Salmonella allow detection of most Salmonella serotypes [12]. Regardless of the detection method used, food samples contaminated by STEC and Salmonella must be enriched in a broth medium that increases the concentration of the pathogen to a detectable level, which is approximately 4 to 5 log10 CFU/mL for most methods. Reaching this effective level can be challenging due to the outgrowth of other naturally present contaminating flora [13]. Numerous methods are used to enhance target pathogen growth such as incubation at restrictive temperatures (eg: 42°C) or inclusion of various antimicrobial compounds [14-16].
In a previous publication we detailed the development of a highly selective enrichment medium for detection and isolation of STEC and Salmonella from ground beef [14]. The media was shown to substantially reduce the complexity of the methods described in the USDA FSIS Microbiological Laboratory Guidebook (MLG) [15]. This was achieved by utilization of selective antimicrobials and inclusion of an efflux pump inhibitor that reduced the growth of background microbiological flora in the food matrix. While the medium was selective, during further validation experiments it was observed to be only effective in beef products rather than all food matrices tested. It was hypothesized that a component inherent in meat was enhancing the growth of STEC and Salmonella. In this report we describe the studies that isolated and characterized the molecular nature of the growth stimulating factor present in ground beef.
Materials and Methods
Approach
The testing of our hypothesis was addressed through the following four experiments:
Experiment 1
Media conditioned with ground beef extract was compared to media containing traditional powdered beef extract supplement as control to verify STEC and Salmonella growth stimulation was directly related to the presence of fresh ground beef.
Experiment 2
Ammonium sulfate precipitation and fractionation, and molecular weight fractionation of ground beef extracts were performed to determine if a specific fraction contained the growth stimulating activity.
Experiment 3
Identification of compound(s) in the active ammonium sulphate fraction was performed by affinity chromatography and preparative SDS-polyacrylamide gel electrophoresis followed by mass spectral analysis of suspect tryptic peptides.
Experiment 4
Commercial sourced biomolecules were obtained and tested for growth stimulating activity to confirm the identity of the active compound(s) Figure 1.
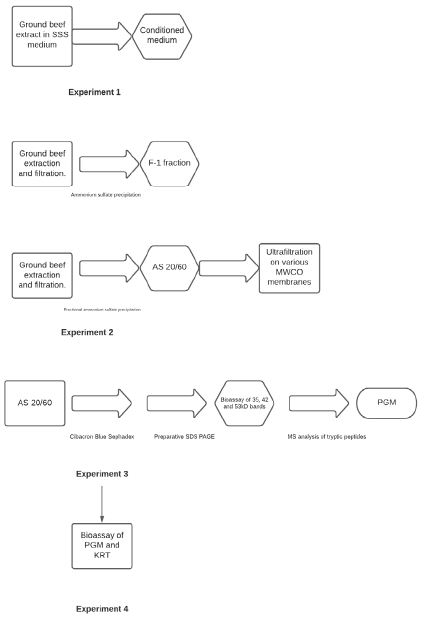
Figure 1: Flow chart of experimental approach.
Media and Media Ingredients
Tryptic Soy Broth (TSB), MI (MUG: methylumbelliferyl-beta-D-galactopyranoside; IBDG: Inoxyl-beta-D-glucuronide) Agar, Brain Heart Infusion (BHI), Buffered Peptone Water, were obtained from Becton Dickinson (Franklin Lakes, NJ). Tryptic Soy Agar (TSA), D-Raffinose, D-Arabinose, Bromocresol Purple, Peptone from casein, D-Xylose were obtained from Sigma-Aldrich (St. Louis, MO). D-Sorbitol was obtained from Fisher Scientific (Hampton, NH). Trehalose was obtained from GoldBio (St. Louis, MO). Bile salt was obtained from Honeywell Fluka (Charlotte, NC). CHROMagarTM STEC and CHROMagarTM SALMONELLA PLUS and were obtained from CHROMagar (Paris, France). The SSS medium, commercially known as PDX-STEC, was prepared according to instructions from the U.S. Patent: 9518283 [17], with the addition of 0.025% (m/v) bromocresol purple. The modified SSS medium or m-SSS medium was prepared by removing sulfanilamide and myricetin from the formulation. Modified tryptic soy broth (mTSB) was prepared by adding 0.15% (w/v) bile salts and 0.0008% (w/v) sodium novobiocin obtained from Sigma Aldrich, Milwaukee, WI. Modified buffered peptone water (mBPWp) was prepared according to the Bacteriological Analytical Manual of the US Food and Drug Administration [18]. Cibacron Blue 3GA was purchased from Polysciences (Warrington, PA).
Bacterial Strains
STEC and Salmonella strains were obtained from the Penn State University E. coli Reference Center in University Park, Pennsylvania, the Center for Disease Control and Prevention in Atlanta, Georgia, the U.S. Meat Animal Research Center (USDA Agricultural Research Services) in Clay Center, Nebraska, the American Type Culture Collection (ATCC) in Manassas, Virginia, and the University of Minnesota Veterinary Diagnostic Laboratory in Saint Paul, Minnesota. Bacterial cultures were maintained as glycerol stock at -20°C and revived in TSB incubated at 37°C overnight before use.
E. coli Growth Stimulating Activity Assays
In Experiments 1 and 2 the growth effects of medium with and without putative stimulating factors were assayed by inoculating 3.0 mL of modified SSS medium or mBPWp with a STEC or Salmonella strain at ~1 CFU/mL. After 7 h of incubation at 37°C, 0.1 mL aliquots of the samples were spread plated on CHROMAGAR™ STEC or CHROMAGAR SALMONELLA PLUS then incubated for 18 hours at 37°C. Bacterial populations were enumerated the following day by colony counts. In Experiments 3 and 4 growth stimulating assays used 1.0 mL portions of SSS medium prepared with purified components or commercial proteins, inoculated with 5-6 CFU/mL of E. coli O157:H7, that were then incubated at 37°C for six hours, after which 0.1 mL was plated onto Chromagar STEC medium. The plates were incubated at 37°C overnight and enumerated the following day.
Time-course experiments were conducted to monitor the growth stimulating activity on STEC and Salmonella in media with and without putative stimulating factors where 100-μL aliquots were withdrawn at 3, 5, and 7 hours and plated onto CHROMAGAR™ STEC or CHROMAGAR SALMONELLA PLUS, incubated and scored as described above.
Assessing Growth Stimulating Factor in Alternate Medium
Wheat kernels (25 g) were placed in stomacher bags then inoculated with ~3 CFU of E. coli O157:H7 and held at room temperature for 20 minutes. Two hundred milliliters of mBPWp, or mBPWp supplemented with 10% (v/v) of the F-1 ammonium sulfate cut (described below) was added to the stomacher bags. The samples were enriched at 42°C for seven hours, and 0.1-mL aliquots were taken at 2-hour intervals and spread onto CHROMAGAR™ STEC plates then incubated overnight at 37°C. Mauve colonies were enumerated.
Conditioning Media
The hypothesized stimulating factor was extracted into SSS medium by incubation of ground beef in ~3:1 v/m ratio where 165 g ground beef (80:20 lean:fat) was suspended in 500 mL SSS medium and stirred for thirty minutes at 10°C. The medium was decanted through a screen in a stomacher bag and filtered through a Celite pad to provide the conditioned medium. The conditioned medium was sterilized by filtration through a 0.22 mm filter.
Extraction Procedure
Ground beef (80:20 lean:fat) was obtained from a local grocery store. Ground beef extracted was prepared by suspending 4:1 v/m in 0.02 M Tris-Cl, pH 7.9, 0.032 M MgCl2, 0.027% w/v Niaproof-4. Extracts were clarified by filtration through course screen in stomacher bags followed by filtration through Celite 545 (Sigma-Aldrich, St. Louis, MO).
Ammonium Sulfate Precipitation and Fractionation
Initial ammonium sulfate fractionation was carried out at 100% saturation to determine if the active component was salt perceptible. To 100 mL extract of the ground beef (see above), 72.9 g of solid ammonium sulfate was added with stirring at 10°C. After 30 minutes, the sample was centrifuged in a MyFuge Mini Centrifuge™, Benchmark Scientific, Edison, NJ, at 6,000 RPM to pellet the precipitate. The supernatant was collected, and the pellet was re-dissolved in 3 mL of 0.01 M Tris-Cl pH 7.8. The resuspended pellet was dialyzed against same buffer to desalt. This initial total fraction was termed “F-1”.
Ammonium sulfate fractionation was carried out by addition of solid ammonium sulfate to obtain 20% and 60% saturation. The protein precipitates obtained at all ammonium sulfate saturation levels were collected by filtration through glass fiber filters and redissolved in a minimum volume of 0.02 M Tris-Cl, pH 7.9. The ammonium sulfate fraction obtained from 20% to 60% saturation was designated “AS-20/60”.
Ammonium sulfate precipitate F-1 and fraction AS-20/60 were prepared in SSS medium to a final concentration of 10% v/v for use in Experiment 2 growth studies with STEC and Salmonella cultures (see above).
Additional Purification Procedures
The F-1 precipitate was further purified using molecular weight cut off filters as follows: The F-1 preparation was ultrafiltered using an Amicon stirred ultrafiltration cell with a 50 and 100 -kD nominal Molecular Weight Cut-off (MWCO) membranes purchased from Sterlitech (Kent, WA). The retentate and filtrate fractions were prepared at 10 % v/v in SSS medium to assayed in Experiment 2 for E. coli growth stimulating activity (above).
The AS-20/60 fraction was further purified on a Cibacron Blue Sephadex column prepared according procedures described by Turner [19], followed by polyacrylamide gel purification according to the procedure of Laemmli [20]. The AS-20/60 fraction was dialyzed into starting buffer, 0.01 M MES, pH 6.1, 0.04 M KCl, 0.001 M dithiothreitol. Then it was loaded onto the column and washed with 5 volumes of starting buffer. The active portion was eluted from the column by washing the column with 3 volumes of starting buffer containing 0.5 M NaCl. The AS-20/60 Sephadex column elutate was loaded into Mini-Protean TGX precast gels (Bio-Rad Laboratories, Hercules CA) for PAGE. Aliquots (10 to 12-uL) having 10 to 100 micrograms protein were applied to the wells and processed according to manufacturer’s instructions. Preparative gels were removed from the gel forms and fixed for 15 minutes in 1 M sodium acetate before negative staining using the zinc-imidazole procedure of Simpson [21]. The visualized bands were excised and minced with a razor blade. Minced bands were suspended overnight in 0.7 mL of 0.02 M Tris-Cl, 0.002 M dithiothreitol pH 7.9 buffer at 4°C. The supernatants obtained were prepared in mBPWp utilizing 10% v/v per assay of the material obtained from the excised protein bands for use in Experiment 3.
Tryptic Digest and Mass Spectrometry Identification of Putative Active Components
The active fractions identified from the polyacrylamide gel purification above were excised from polyacrylamide gel slab suspended in 2.0 mL 0.02 M Tris-Cl pH 7.8. The samples were submitted to the Center for Proteomics Mass Spectroscopy facility at the University of Minnesota. The samples were digested with Trypsin and subjected to mass spectroscopic analysis according to procedures previously published [22].
Putative Growth Factor Preparation
Rabbit muscle Phosphoglucomutase (PGM) was purchased from Sigma-Aldrich (Milwaukee, WI). Keratin was purchased from Fitzgerald Industries International (Acton, MA). PGM was diluted to 1 mg/mL in 0.02 M Tris-Cl pH 7.0, 0.001 M dithiothreitol (Sigma Aldrich, Milwaukee, WI). The PGM was prepared in mBPWp at 50 mg/mL and 100 mg/mL to test growth stimulating activity. Keratin was dissolved in 0.02 M Tris-Cl pH 7.0, 0.001 M dithiothreitol at 1 mg/mL and used at 50- and 100 µg/mL in mBPWp for growth stimulating assays.
Statistical Analysis
All cell count assays were performed in triplicate except where specifically mentioned above. Colony Forming Units (CFU) per mL were log transformed for analysis. Mean log10 CFU/mL and standard deviations were calculated using the AVERAGE and SDIFF functions in Microsoft Excel. Unpaired t-tests to identify significantly different means were performed using GraphPad Prizm quick calcs (www.graphpad.com/quickcalcs/ttest) with significant difference set at 0.05.
Results
Experiment 1
The initial experiment used conditioned SSS medium containing ground beef extract and compared that to SSS containing traditional powdered beef extract supplement (as control) to verify STEC and Salmonella growth stimulation was directly related to the ground beef extract. The increases in 7 h populations of roughly 50-fold for E. coli O157:H7 and ~300-fold for STEC-O45 suggested a growth stimulating compound was provided by extracts from fresh ground beef, but not any compounds present in powdered beef extract (Table 1).
Table 1: Effect of Supplementing Media on STECa Growthb.
|
E. coli Strain |
Medium Supplementationd | ||
| None | Beef Extract Powere |
Ground Beef Conditionedf |
|
|
O157:H7 |
<LODg | 2.1 ± 0.03 | 3.7 ± 0.024h |
| STEC-O45 | <LOD | 0.4 ± 0.12 |
2.5 ± 0.018h |
aSTEC are Shiga toxin-producing E. coli.
bValues represent mean Log10 CFU/mL ± standard deviation attained by a 1-3 CFU/mL of each strain following 7 h incubation at 42°C.
dThe highly selective SSS medium was used with supplements shown.
eBeef Extract Powder was supplemented at 5% (w/v) into SSS broth.
fConditioning of media was accomplished by incubation of ground beef in SSS media (3:1 v/m ratio) stirred 30 m at 10°C, then clarified by screening and filtering before sterilized using a 0.22 mm filter.
gValue below the level of detection (LOD) of 0.0 Log10 CFU/mL.
hThe difference between the two supplements is significantly different (P< 0.05).
Experiment 2
We commenced to partially purify the putative growth stimulating factor from crude ground beef extract by ammonium sulfate precipitation. The total precipitate, referred to as F-1, was used to supplement SSS media and compared to control SSS for growth stimulating activity after 7 h of incubation. STEC (O157, O111, and O45) at ~1 CFU/mL were demonstrated to reach concentrations of ~3 logs CFU greater over controls that lacked the growth stimulating factor supplied by the F-1 preparation (Table 2). The results further showed that using the more concentrated F-1 preparation provided a factor of ~100-fold (2 log) more colonies than the simply conditioned media in Experiment 1 (Table 1). Thus, demonstrating the growth stimulating factor was enriched in the F-1 preparation, and its activity was concentration dependent when examined under similar incubation conditions.
Table 2: Effect of Ammonium Sulfate Precipitate Fraction 1 (F-1) on STECa growthb.
|
E. coli Strain |
Medium Supplementationd | |
| None |
F-1 Precipitatee |
|
|
O157:H7 |
2.2 ± 0.07 | >4.0f h |
| STEC-O111 | 2.1 ± 0.05 |
>4.0f h |
|
STEC-O45 |
<LODg |
3.7 ± 0.03h |
aSTEC are Shiga toxin-producing E. coli.
bValues represent mean Log10 ± standard deviation CFU/mL attained by a 1-3 CFU/mL of each strain following 7h incubation at 42°C.
dThe highly selective SSS medium was used with supplements shown.
eThe F-1 Precipitate was a saturated ammonium sulphate precipitation from a ground beef suspension, that was used at X% (w/v) in SSS broth.
fThe upper limit of resolution in the colony counting assay was limited to 4.0 Log10 CFU/mL with these sample results being too numerous to count.
gValue below the level of detection (LOD) of 0.0 Log10 CFU/mL.
hThe difference between the two supplements is significantly different (P< 0.05).
The activity of the F-1 precipitate was examined over time on STEC and Salmonella. Growth Time points were taken every two hours and compared to control cultures. Growth was observed to be accelerated for three different STEC serotypes in the presence of SSS medium supplemented with 10% v/v F-1 preparation (Figure 1). STEC-O157:H7, -O111, and -O121 populations entered log phase growth in the presence of 10% v/v F-1 at a time point at where matched controls were still in lag phase growth (Figure 2). In analogous experiments using different Salmonella serotypes (Newport, Heidelberg, and Tennessee) similar activity of 10% v/v F-1 in SSS media was observed (Figure 3).
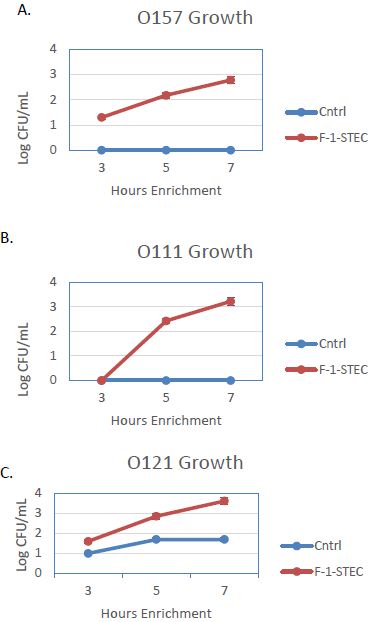
Figure 2: Growth curves for STEC O157:H7 (A), STEC-O111 (B), and STEC-O121 (C) in SSS media (Cntrl; blue – diamond) and in SSS media containing 10% v/v F-1 preparation from ammonium sulfate precipitation of ground beef extract (F-1-STEC; orange – square) measured at 3, 5 and 7 h post inoculation.
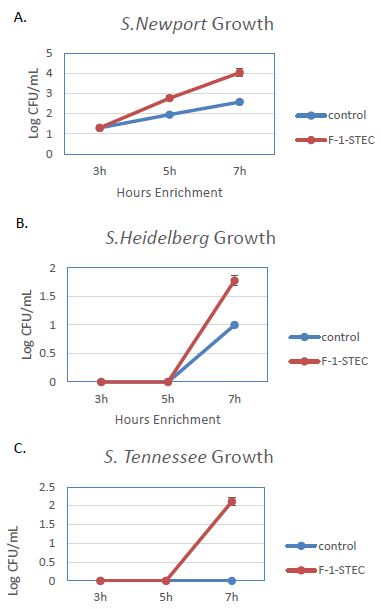
Figure 3: Growth curves for Salmonella newport (A), Salmonella heidelberg (B), and Salmonella tennessee (C) in modified SSS medium (Cntrl; blue) and in modified SSS medium containing 10% v/v F-1 fraction from ammonium sulfate precipitation of ground beef extract (F-1-STEC; orange) measured at 3, 5 and 7 h post inoculation.
Having determined that the F-1 precipitate influenced STEC and Salmonella growth, the ammonium sulphate precipitation was refined by fractionation to identify where the activity was most concentrated. The precipitate formed by the 20 to 60% ammonium sulphate fraction (AS-20/60) was found to possess >90% of the stimulating activity (Table 3). Then to further characterize the growth stimulating factor, its apparent molecular weight was estimated by ultrafiltration of the AS-20/60 fraction with 50- and 100 -kD nominal Molecular Weight Cut-off (MWCO) membranes. When the filtrate and retentate of each were tested for E. coli O157:H7 growth stimulation, the 50-kD retentate and larger molecular weight fractions were found to be most active (Table 4). The molecular weight of the growth stimulating factor was considered to be >50,000 and <100,000 molecular weight for Experiment 3.
Table 3: Growth Stimulation of E. coli O157:H7a by Ammonium Sulfate Fractionsb.
| Ammonium Sulphate Fractionc | |||
|
Controld |
0-20% | 20-60% |
60-90% |
| <LODe | 0.8 ± 0.10 | 2.4 ± 0.05f |
<LOD |
aValues represent mean Log10 ± standard deviation CFU/mL attained by a 1-3 CFU/mL of E. coli O157:H7 following 7h incubation at 42°C.
bThe highly selective SSS medium was used and supplemented with the ammonium sulphate fractions shown
cEach fraction was used at X% (w/v) in the SSS media broth.
dControl was non-supplemented SSS media.
eValue below the level of detection (LOD) of 0.0 Log10 CFU/mL.
fThe difference between the values for this ammonium sulphate fraction compared to the control is significantly different (P<0.05).
Table 4: Growth Stimulation of E. coli O157:H7a by ultrafiltration fractions of AS-20/60b.
|
50 kD MWCOc |
100 kD MWCO |
|||
|
Controld |
Filtratee | Retentatef | Filtrate | Retentate |
| <LODg | <LOD | 4.2 ± 0.06h | 4.2 ± 0.04h |
4.3 ± 0.02h |
aValues represent mean Log10 ± standard deviation CFU/mL attained by a 1-3 CFU/mL of E. coli O157:H7 following 7h incubation at 42°C.
bThe highly selective SSS medium was used and supplemented with the ultrafiltrate fractions shown. Each fraction was used at X% (v/v) in the SSS media broth.
cMWCO = Molecular Weight Cut Off.
dControl was non-supplemented SSS media.
eFiltrate was the fraction that passed through the MWCO membrane, and is presumed to contain proteins lower than the MWCO.
fRetentate was the fraction that did not pass through the MWCO, and is presumed to contain proteins greater than the MWCO.
gValue below the level of detection (LOD) of 0.0 Log10 CFU/mL.
hThe difference between the value for this filtrate or retentate compared to the control is significantly different (P<0.05).
Experiment 3
Candidate compounds responsible for growth stimulation were identified and characterized. To obtain higher purity preparation than ammonium sulfate fractionation we subjected the AS-20/60 fraction to purification on CibaCron Blue Sephadex that efficiently bound the growth stimulating substance. The growth stimulating activity was eluted from the affinity matrix and was further resolved by SDS PAGE. This resulted in several SDS PAGE bands predominantly in the 30- to 60- kD range with the most intensely staining bands at approximately 35-, 42-, and 52- kD. Similar suspect molecular weight proteins were observed on negatively stained non-denaturing PAGE. PAGE bands of approximately 20-, 35-, 42- and 52- kD were excised and tested for E. coli O157:H7 growth stimulating activity in mBPWp (Table 5). PAGE bands of approximately 20, 35, and 52-kD increased E. coli O157:H7 growth by 0.5 to 0.8 Log10, while the prominent 42 kD band had no activity. The bands corresponding to the 35-, 42-, and 52-kD proteins were submitted for mass spectroscopic analysis of their tryptic digests.
Table 5: Growth Stimulation of E. coli O157:H7a by PAGE Gel Band Preparationsb.
|
PAGE Gel Band Preparationc |
|||||
|
Controld |
20 kD | 35 kD | 42 kD | 52 kD | |
| 2.7 ± 0.01 | 3.2 ± 0.01e | 3.1 ± 0.01e | 2.6 ± 0.04e |
3.3 ± 0.04e |
|
aValues represent mean Log10 ± standard deviation CFU/mL attained by a 1-3 CFU/mL of E. coli O157:H7 following 7h incubation at 42°C.
bProminent non-denaturing PAGE gel bands were excised, minced and extracted to determine which possessed growth stimulating activity.
cEach preparation was used at 10% (v/v) in mBPWp broth.
dControl was non-supplemented mBPWp.
eThe difference between the value for this band preparation compared to the control is significantly different (P< 0.05).
The resulting mass spectroscopy data (Tables S1, S2, and S3) showed a large percentage of the spectrum in the 35-kD protein band corresponded to keratin type proteins, KRT 2, KRT10, KRT 14 and KRT 9; whereas the mass spectrum data from the excised 52-kD band was mostly devoid of any keratin proteins. The one protein appearing in the MS analysis of both the 35-kD and 52-kD protein bands was phosphoglucomutase. The spectrum of the 42-kD protein, the inactive band, was primarily creatine phosphokinase.
Experiment 4
The identity of the active compound stimulating the growth of STEC and Salmonella was confirmed with commercially sourced biomolecules. The candidate proteins keratin and phosphoglucomutase were obtained and tested at two concentrations for growth stimulating activity to confirm the identity of the active compound (Table 6). This demonstrated that the putative growth stimulating factor comprises phosphoglucomutase as suggested by the mass spectroscopy results of the excised active PAGE bands. The commercially sourced PGM demonstrated concentration dependent stimulation of E. coli O157:H7 growth as characterized in the crude F-1 and AS-20/60 fractions. Further, while the protein keratin was found to be abundant in the mass spectroscopy analysis, it clearly had no growth stimulating activity and demonstrated a mild inhibitory activity. See Figure 1 depicting the experimental approach overview.
Table 6: Growth Stimulation of E. coli O157:H7a by candidate proteins obtained from commercial sourcesb.
|
PGM (ug/mL) |
KRT (ug/mL) |
|||
|
Controlc |
50 | 100 | 50 | 100 |
| 1.5 ± 0.10 | 2.1 ± 0.11e | 2.4 ± 0.03e | <LODd |
1.0 ± 0.12e |
aValues represent mean Log10 ± standard deviation CFU/mL attained by a 1-3 CFU/mL of E. coli O157:H7 following 7h incubation at 42°C.
bCommercially available rabbit muscle phosphoglucomutase (PGM) and keratin (KRT) were supplemented at two concentrations into SSS media broth.
cControl was non-supplemented SSS media broth.
dValue below the level of detection (LOD) of 0.0 Log10 CFU/mL.
eThe difference between the value for this this protein at this concentration compared to the control is significantly different (P< 0.05).
We anticipated that the use of PGM in an enrichment medium would decrease the time to detection for STEC and Salmonella, especially for samples that are not beef or meat. To examine this, an enrichment of wheat inoculated with E. coli O157:H7 was carried out in mBPWp and mBPWp supplemented with the PGM containing F-1 fraction. The growth curves from this demonstration showed that the PGM present in the F-1 fraction stimulated the growth of the E. coli O157:H7 over the control and would lead to more rapid detection (Figure 4).
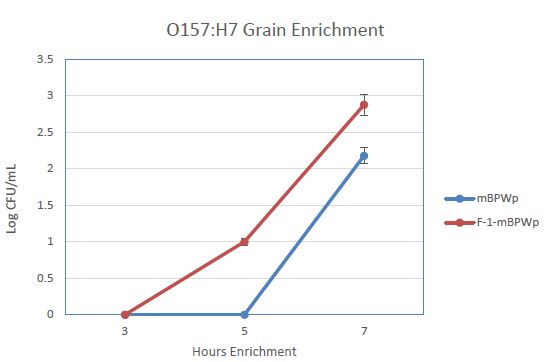
Figure 4: Effect of Phoshoglucomutase (PGM) containing F-1 fraction on the growth of E. coli O157:H7 wheat samples. Medium (modified Buffered Peptone Water with pyruvate; mBPWp) supplemented with PGM (10% F-1; orange) compared to control using mBPWp (blue) was measured over 7 h of incubation at 42°C.
Discussion
We entered into these experiments because we observed that inoculated spinach enrichments using the selective medium, SSS, at seven hours enrichment no detectable STEC colonies were found on plating medium while in comparable meat enrichments there were easily detectable numbers of STEC colonies. This suggested that there was either an active component provided by beef, or that there was an inhibitory compound supplied by the spinach. It could also be argued that since SSS media uses a number of components that inhibit background microflora growth, the beef was releasing a substance that negated the selectivity of the SSS media. However, since SSS media has been well characterized and defined for use in beef, and some control STEC and Salmonella strains grow slowly as a pure culture in SSS media [14], we decided to test the hypothesis that there was a component inherent in meat that was enhancing the growth of STEC and Salmonella.
We proceeded to isolate and characterize the growth stimulating activity extractable from beef tissues. Conditioned medium was shown to have 50-fold greater growth of E. coli O157:H7 than cultures of non-supplemented medium; and the same conditioned medium exhibited over 300-fold greater growth than the non-supplemented medium. The active component was shown to be precipitable in saturated ammonium sulfate solution permitting partial purification of the protein. Greater than 90% of the growth stimulating activity could be obtained by taking the 20%-60% ammonium sulfate saturation interval. As with the STEC growth stimulation effect, we demonstrated that the protein stimulates the growth of several Salmonella serotypes.
One of the earliest media used to cultivate bacteria was one that contained an infusion of meat [23]. Beef or meat extract has been a commonly used nutrient source in microbiology ever since. Current Beef Heart Infusion (BHI) or beef extract powders are intended to replace the classical aqueous infusions of meat in culture media. Typical preparations of beef extract are a mixture of peptides, amino acids, nucleotides, organic acids, minerals and some vitamins. Manufacture of BHI and beef extract powders employ techniques that can hydrolyze or denature the activity of PGM when it is present. We suspect that this is why these media supplements lack the activity we identified in our experiments.
Fratamico et al. [24] demonstrated that ground beef extracts activated genes associated with E. coli survival, particularly those associated with acid shock exposure. Although their findings did not hint at the apparent growth stimulating effects of a ground beef extractable protein. Harhay et al. [16] found that ground beef enrichments supported more rapid growth of Salmonella than parallel control enrichments in mTSB. The authors were focused on the impact of this observation on the prediction model accuracy rather than theorize on the reasons for the reduction in doubling time for both the slow and fast-growing Salmonella strains in media containing ground beef versus only mTSB.
After obtaining a more highly purified preparation from affinity chromatography we noted that the predominant bands, 35 kD, 42 kD and 52 kD SDS PAGE bands were within the 50 kD-100 kD molecular weight range from the ultrafiltration experiments, assuming that the smaller proteins might exist as dimers. To verify that these proteins were growth stimulatory we isolated them from PAGE gels under minimally denaturing conditions. The assay for E. coli growth stimulating activity identified three PAGE bands, two of which were submitted for mass spectroscopy along with one inactive band to determine their identities. Common proteins were identified by the MS analysis in the active bands that were absent from the inactive band. Keratin was initially considered to be the candidate protein; however, it was abundant and appeared in both the active and inactive band MS analyses. The 42 kD band was rich in creatine phosphokinase which has been reported to be growth inhibitory towards E. coli [25]. The single protein found in the MS spectrum of both active bands (the 35 kD and 52 kD PAGE bands) analyzed was Phosphoglucomutase (PGM). Purchased commercial rabbit muscle PGM was shown to exhibit appreciable E. coli growth stimulating activity while commercial keratin was devoid of growth stimulating activity.
The enzyme, PGM (E.C. 5.4.2.2), plays a central role in intermediary metabolism of glucose by inter-converting glucose1-phosphate with glucose-6-phosphate allowing the latter to enter the glycolytic pathway to generate cellular energy [26]. PGM mutants in E. coli are defective in their ability to utilize galactose as a carbon source since it cannot be converted to glucose 6-phosphate from glucose-1-phosphate, which ultimately generates energy via the glycolytic pathway [27]. Patterson et al. reported that PGM deletion mutants in Salmonella serotype Typhimurium were defective in O-antigen synthesis; were more susceptible to antimicrobial peptides and were less able to survive in infected mice than the wild type strain [28]. The authors concluded that PGM played a critical role in imparting fitness and adaptability to Salmonella Typhimurium. These findings appear to comport with our observations that PGM stabilizes the growth of STEC and Salmonella in a highly selective medium.
STEC and Salmonella commandeer the catecholamines in the gastrointestinal tract to stimulate their growth through induction of an autoinducer molecule [29,30]. An example of bacterial symbionts assimilating host enzymes to stimulate their growth is novel. Since the structure and function of PGM is evolutionarily conserved [31], it is possible that bacterial assimilation may be more readily achieved. The ability to commandeer host enzymes for survival and growth gives bacterial symbionts remarkable environmental adaptability. A recent in-silico study [32,33] examined numerous host pathogen protein interactions and implicated several bacterial enzymes and proteins in the pathogenesis process, however nothing quite like the assimilation of particular host proteins to facilitate pathogen adaptation and survival in the host environment.
Conclusion
In conclusion, we recognized and identified PGM as a growth stimulating substance in ground beef extracts. While much of this study was conducted utilizing bovine tissues, PGM is present at varying levels in all eukaryotic organisms. Its greater activity in meat samples compared to spinach may be due to the cell wall of plants prohibiting its release into the enrichment medium. Although these results need further development current data indicate that PGM can serve as a supplement in numerous enrichment media to improve pathogen detection.
Acknowledgment
The authors wish to acknowledge Paradigm Diagnostics for funding this project. We also wish to acknowledge the helpful input of the scientists at the University of Minnesota Center for Proteomics. We also thank Dr Tommy Wheeler for critical review of this manuscript and Jody Gallagher for administrative support. USDA is an equal opportunity provider and employer. Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
References
- Mckay B. Aurora Packing Company, Inc. Recalls Beef Products Due to Possible coli O157:H7 Contamination.
- Pfaeffle V. FSIS Issues Public Health Alert for Ready-to-Eat Meat and Poultry Products Containing FDA-Regulated Onions that have been Recalled due to Possible Salmonella Newport Contamination.
- Okonta C. FSIS Issues Public Health Alert for Raw Beef Ravioli Products Due to Possible coli O157:H7 Contamination.
- Hirschmugl J. Meijer Recalls Whole Cantaloupes and Select Cut Cantaloupe Trays Due to Potential Health Risk.
- Seneca Recalls Cinnamon Apple Chips Because of Possible Health Risk.
- Prima® Wawona Recalls Bulk/Loose and Bagged Peaches Due to Possible Salmonella Risk.
- King Arthur Flour Updates Three Lot Codes of Voluntarily Recalled Unbleached All-Purpose Flour (5 lb).
- Centers for Disease Control and Prevention (CDC) (2007) Multistate outbreak of Salmonella serotype Tennessee infections associated with peanut butter–United States, 2006-2007. MMWR Morb Mortal Wkly Rep 56: 521-524.
- Bosilevac JM, Kalchayanand N, Schmidt JW, Shackelford SD, Wheeler TL, et al. (2010) Inoculation of beef with low concentrations of Escherichia coli O157:H7 and examination of factors that interfere with its detection by culture isolation and rapid methods. J Food Prot 73: 2180-2188. [crossref]
- Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, et al. (2005) Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. The Journal of Infectious Diseases 192: 1422-1429. [crossref]
- Ceuppens S, Li D, Uyttendaele M, Renault P, Ross P, et al. (2014) Molecular Methods in Food Safety Microbiology: Interpretation and Implications of Nucleic Acid Detection. Comprehensive Reviews in Food Science and Food Safety 13: 551-577.
- Sunar NM, EI Stentiford, DI Stewart, LA Fletcher (2010) Molecular techniques to characterize the invA genes of Salmonella spp. for pathogen inactivation study in composting.
- Eggers J, JM Feirtag, AD Olstein, JM Bosilevac (2018) A Novel Selective Medium for Simultaneous Enrichment of Shiga Toxin-Producing Escherichia coli and Salmonella in Ground Beef. J Food Prot 81: 1252-1257. [crossref]
- S. Department of Agriculture, Food Safety Inspection Service. 2014. Microbiology laboratory guidebook. Method no. 5B.05.
- Harhay DM, Weinroth MD, Bono JL, Harhay GP, Bosilevac JM (2020) Rapid estimation of Salmonella enterica contamination level in ground beef -Application of the time-to-positivity method using a combination of molecular detection and direct plating. Food Microb 93: 103615-103623.
- Olstein AD (2016) Selective enrichment media and uses there of US Patent 9,518,283.
- FM Huntoon (1918) “Hormone” Medium: A Simple Medium Employable as a Substitute for Serum Medium. J Infect Dis 23: 169-172.
- Turner AJ (1979) A Simple and Colorful Procedure to Demonstrate the Principles of Affinity Chromatography. Ed 7: 60-62.
- Leammli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685. [crossref]
- Simpson RJ (2007) Zinc/Imidazole Procedure for Visualization of Proteins in Gels by Negative Staining. Cold Spring Harbor Protocols [crossref]
- Thu YM, Van Riper SK, Higgins L, Zhang T, Becker JR, et al. (2016) Slx5/Slx8 Promotes Replication Stress Tolerance by Facilitating Mitotic Progression. Cell Rep 15: 1254-1265. [crossref]
- Snyder TR, Boktor SW, M’ikanatha NM (2019) Salmonellosis Outbreaks by Food Vehicle, Serotype, Season, and Geographical Location, United States, 1998 to 2015. Journal of Food Protection 82: 1191-1199. [crossref]
- Fratamico PM, Wang S, Yan X, Zhang W, Li Y (2011) Differential gene expression of coli O157:H7 in ground beef extract compared to tryptic soy broth. J Food Sci 76: M79-87. [crossref]
- An F, Fan N, Zhang S (2009) Creatine kinase is a bacteriostatic factor with a lectin-like activity. Immun 46: 2666-2670. [crossref]
- 540, Chap. 9 in Biochemistry, The Chemical Reaction of Living Cells. David Metzler ed. Academic Press. 1977.
- Adhya S, Schwartz M (1971) Phosphoglucomutase mutants of Escherichia coli K-12. J Bacteriology 108: 621-626. [crossref]
- Patterson GK, Cone DB, Peters SE, Maskell DJ (2009) The enzyme Phosphoglucomutase (PGM) is required by Salmonella enterica serovar Typhimurium for O-antigen production, resistance to antimicrobial peptides and in vivo fitness. Microbiology 155: 3403-3410. [crossref]
- Lyte M, Frank CD, Green BT (1996) Production of an autoinducer of growth by norepinephrine cultured Escherichia coli O157:H7. FEMS Microbiology Letters 139: 155-159. [crossref]
- Pullinger GD, Carnell SC, Sharaff FF, van Dieman PM, Dziva F, et al. (2010) Norepinephrine Augments Salmonella enterica-Induced Enteritis in a Manner Associated with Increased Net Replication but Independent of the Putative Adrenergic Sensor Kinases QseC and QseE. Infection and Immunity 78: 372-380. [crossref]
- Mehra-Chaudhary R, Mick J, Tanner JJ, Henzle MT, Beamer LJ (2011) Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens. Proteins 79: 1215-1229. [crossref]
- Bose T, Venkatesh KV, Mande, SS (2019) Investigating host-bacterial interactions among enteric pathogens. BMC Genomics 20: 1022. [crossref]
- S. Food and Drug Administration. Peter Feng and Karen Jinneman BAM: Diarrheagenic Escherichia coli. 2017.
- Turro NJ, Lei X, Ananthapadmanabhan KP, Aronson M (1995) Spectroscopic probe analysis of protein–surfactant interactions: The BSA/SDS system. Langmuir 11: 2525-2533.
Supplementary Files
Table S1: Scaffold file of mass spectrum of 52-kD excised protein band.
| # | Identified Proteins (61) | Alternate ID | Molecular Weight | Percentage Total Spectrum_ |
| 1 | Serum albumin OS=Bos taurus OX=9913 GN=ALB PE=4 SV=1 | ALB | 69 kDa | 4.90% |
| 2 | Methanethiol oxidase OS=Bos taurus OX=9913 GN=SELENBP1 PE=1 SV=1 | SELENBP1 | 53 kDa | 2.00% |
| 3 | Glucose-6-phosphate isomerase OS=Bos taurus OX=9913 GN=GPI PE=1 SV=1 | GPI | 64 kDa | 0.99% |
| 4 | Retinal dehydrogenase 1 OS=Bos taurus OX=9913 GN=ALDH1A1 PE=1 SV=3 | ALDH1A1 | 55 kDa | 0.82% |
| 5 | Aldehyde dehydrogenase, mitochondrial OS=Bos taurus OX=9913 GN=ALDH2 PE=1 SV=2 | ALDH2 | 57 kDa | 0.66% |
| 6 | Trypsin OS=Sus scrofa OX=9823 PE=1 SV=1 | 24 kDa | 0.58% | |
| 7 | Thioredoxin reductase 1, cytoplasmic OS=Bos taurus OX=9913 GN=TXNRD1 PE=2 SV=3 | TXNRD1 | 55 kDa | 0.39% |
| 8 | Keratin, type II cytoskeletal 1 OS=Homo sapiens OX=9606 GN=KRT1 PE=1 SV=6 | KRT1 | 66 kDa | 0.37% |
| 9 | PGM5 protein OS=Bos taurus OX=9913 GN=PGM5 PE=2 SV=1 | PGM5 | 62 kDa | 0.33% |
| 10 | Glutathione S-transferase P OS=Bos taurus OX=9913 GN=GSTP1 PE=1 SV=2 | GSTP1 | 24 kDa | 0.41% |
| 11 | Alpha-1-antiproteinase OS=Bos taurus OX=9913 GN=SERPINA1 PE=1 SV=1 | SERPINA1 | 46 kDa | 0.37% |
| 12 | Cytosol aminopeptidase OS=Bos taurus OX=9913 GN=LAP3 PE=1 SV=3 | LAP3 | 56 kDa | 0.33% |
| 13 | Cluster of Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 (P13645) | KRT10 | 59 kDa | 0.35% |
| 13.1 | Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 | KRT10 | 59 kDa | 0.27% |
| 13.2 | Keratin, type I cytoskeletal 14 OS=Bos taurus OX=9913 GN=KRT14 PE=1 SV=1 | KRT14 | 50 kDa | 0.12% |
| 13.3 | IF rod domain-containing protein OS=Bos taurus OX=9913 GN=KRT13 PE=3 SV=1 | KRT13 | 47 kDa | 0.10% |
| 13.4 | Keratin, type I cytoskeletal 19 OS=Bos taurus OX=9913 GN=KRT19 PE=2 SV=1 | KRT19 | 44 kDa | 0 |
| 14 | Hemopexin OS=Bos taurus OX=9913 GN=HPX PE=2 SV=1 | HPX | 52 kDa | 0.35% |
| 15 | Keratin, type I cytoskeletal 9 OS=Homo sapiens OX=9606 GN=KRT9 PE=1 SV=3 | KRT9 | 62 kDa | 0.27% |
| 16 | Uncharacterized protein OS=Bos taurus OX=9913 PE=1 SV=1 | 35 kDa | 0.27% | |
| 17 | Alanine aminotransferase 1 OS=Bos taurus OX=9913 GN=GPT PE=4 SV=1 | GPT | 88 kDa | 0.25% |
| 18 | Dihydrolipoyl dehydrogenase OS=Bos taurus OX=9913 GN=DLD PE=1 SV=2 | DLD | 54 kDa | 0.25% |
| 19 | Keratin, type II cytoskeletal 2 epidermal OS=Homo sapiens OX=9606 GN=KRT2 PE=1 SV=2 | KRT2 | 65 kDa | 0.23% |
| 20 | Creatine kinase B-type OS=Bos taurus OX=9913 GN=CKB PE=1 SV=1 | CKB | 43 kDa | 0.25% |
| 21 | Glutathione reductase OS=Bos taurus OX=9913 GN=GSR PE=3 SV=3 | GSR | 56 kDa | 0.23% |
| 22 | Rab GDP dissociation inhibitor alpha OS=Bos taurus OX=9913 GN=GDI1 PE=1 SV=1 | GDI1 | 51 kDa | 0.21% |
| 23 | Serotransferrin OS=Bos taurus OX=9913 GN=TF PE=1 SV=2 | TF | 78 kDa | 0.23% |
| 24 | WD repeat-containing protein 1 OS=Bos taurus OX=9913 GN=WDR1 PE=4 SV=2 | WDR1 | 66 kDa | 0.18% |
| 25 | Phosphoglucomutase-1 OS=Bos taurus OX=9913 GN=PGM1 PE=3 SV=1 | PGM1 | 62 kDa | 0.16% |
| 26 | Lymphocyte cytosolic protein 1 OS=Bos taurus OX=9913 GN=LCP1 PE=1 SV=1 | LCP1 | 70 kDa | 0.19% |
| 27 | Peptidase D OS=Bos taurus OX=9913 GN=PEPD PE=3 SV=1 | PEPD | 55 kDa | 0.18% |
| 28 | Phosphoglucomutase 2 OS=Bos taurus OX=9913 GN=PGM2 PE=1 SV=1 | PGM2 | 67 kDa | 0.16% |
| 29 | Thioredoxin reductase 2, mitochondrial OS=Bos taurus OX=9913 GN=TXNRD2 PE=1 SV=2 | TXNRD2 | 55 kDa | 0.18% |
| 30 | Serpin A3-3 OS=Bos taurus OX=9913 GN=SERPINA3-3 PE=1 SV=1 | SERPINA3-3 | 46 kDa | 0.12% |
| 31 | Hydroxyacyl-CoA dehydrogenase OS=Bos taurus OX=9913 GN=HADH PE=4 SV=1 | HADH | 34 kDa | 0.14% |
| 32 | Carboxypeptidase B2 OS=Bos taurus OX=9913 GN=CPB2 PE=4 SV=1 | CPB2 | 44 kDa | 0.08% |
| 33 | Glucosylceramidase beta 3 OS=Bos taurus OX=9913 GN=GBA3 PE=3 SV=3 | GBA3 | 54 kDa | 0.08% |
| 34 | Cluster of SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC112445741 PE=3 SV=1 (A0A3Q1MGZ6) | LOC112445741 | 45 kDa | 0.08% |
| 34.1 | SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC112445741 PE=3 SV=1 | LOC112445741 | 45 kDa | 0.06% |
| 34.2 | Serpin A3-7 OS=Bos taurus OX=9913 GN=SERPINA3-7 PE=1 SV=1 | SERPINA3-7 | 47 kDa | 0.06% |
| 35 | Alpha-enolase OS=Bos taurus OX=9913 GN=ENO1 PE=3 SV=1 | ENO1 | 45 kDa | 0.08% |
| 36 | Inter-alpha-trypsin inhibitor heavy chain H4 OS=Bos taurus OX=9913 GN=ITIH4 PE=4 SV=1 | ITIH4 | 101 kDa | 0.08% |
| 37 | Aldo-keto reductase family 1 member A1 OS=Bos taurus OX=9913 GN=AKR1A1 PE=2 SV=1 | AKR1A1 | 37 kDa | 0.08% |
| 38 | Bifunctional purine biosynthesis protein ATIC OS=Bos taurus OX=9913 GN=ATIC PE=2 SV=1 | ATIC | 64 kDa | 0.06% |
| 39 | Uncharacterized protein OS=Bos taurus OX=9913 PE=1 SV=1 | 40 kDa | 0.10% | |
| 40 | Aldo-keto reductase family 1 member B1 OS=Bos taurus OX=9913 GN=AKR1B1 PE=1 SV=2 | AKR1B1 | 36 kDa | 0.06% |
| 41 | Ceruloplasmin OS=Bos taurus OX=9913 GN=CP PE=1 SV=1 | CP | 116 kDa | 0.06% |
| 42 | Superoxide dismutase [Cu-Zn] OS=Bos taurus OX=9913 GN=SOD3 PE=1 SV=1 | SOD3 | 27 kDa | 0.04% |
| 43 | Alpha-1B-glycoprotein OS=Bos taurus OX=9913 GN=A1BG PE=1 SV=1 | A1BG | 62 kDa | 0.04% |
| 44 | Transthyretin OS=Bos taurus OX=9913 GN=TTR PE=3 SV=1 | TTR | 20 kDa | 0.04% |
| 45 | Cluster of SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC511695 PE=3 SV=1 (A0A3Q1LY36) | LOC511695 | 45 kDa | 0.04% |
| 45.1 | SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC511695 PE=3 SV=1 | LOC511695 | 45 kDa | 0.04% |
| 45.2 | SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC511106 PE=3 SV=3 | LOC511106 | 44 kDa | 0.02% |
| 46 | Alpha-aminoadipic semialdehyde dehydrogenase OS=Bos taurus OX=9913 GN=ALDH7A1 PE=3 SV=1 | ALDH7A1 | 59 kDa | 0.04% |
| 47 | Cluster of Glutathione S-transferase OS=Bos taurus OX=9913 GN=GSTM3 PE=1 SV=1 (A0A3Q1LSN6) | GSTM3 | 28 kDa | 0.04% |
| 47.1 | Glutathione S-transferase OS=Bos taurus OX=9913 GN=GSTM3 PE=1 SV=1 | GSTM3 | 28 kDa | 0.04% |
| 47.2 | Glutathione S-transferase OS=Bos taurus OX=9913 GN=GSTM2 PE=3 SV=1 | GSTM2 | 26 kDa | 0 |
| 48 | Transgelin OS=Bos taurus OX=9913 GN=TAGLN PE=1 SV=4 | TAGLN | 23 kDa | 0.04% |
| 49 | SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC112445470 PE=3 SV=1 | LOC112445470 | 28 kDa | 0.04% |
| 50 | EMAP like 2 OS=Bos taurus OX=9913 GN=EML2 PE=4 SV=1 | EML2 | 98 kDa | 0.04% |
| 51 | Fascin OS=Bos taurus OX=9913 GN=FSCN1 PE=1 SV=1 | FSCN1 | 55 kDa | 0.04% |
| 52 | KRT5 protein OS=Bos taurus OX=9913 GN=KRT5 PE=2 SV=1 | KRT5 | 63 kDa | 0.06% |
| 53 | Fructose-bisphosphate aldolase OS=Bos taurus OX=9913 GN=ALDOA PE=1 SV=1 | ALDOA | 39 kDa | 0.04% |
| 54 | Keratin, type II cytoskeletal 79 OS=Bos taurus OX=9913 GN=KRT79 PE=3 SV=1 | KRT79 | 57 kDa | 0.04% |
| 55 | IF rod domain-containing protein OS=Bos taurus OX=9913 GN=KRT6A PE=3 SV=1 | KRT6A | 61 kDa | 0.04% |
Table S2: Scaffold file of mass spectrum of 35-kD excised protein band.
| # | Identified Proteins (55) | Alternate ID | Molecular Weight | Percentage of
Total Spectrum |
| 1 | Cluster of Keratin, type II cytoskeletal 2 epidermal OS=Homo sapiens OX=9606 GN=KRT2 PE=1 SV=2 (P35908) | KRT2 | 65 kDa | 2.90% |
| 1.1 | Keratin, type II cytoskeletal 2 epidermal OS=Homo sapiens OX=9606 GN=KRT2 PE=1 SV=2 | KRT2 | 65 kDa | 1.70% |
| 1.2 | KRT5 protein OS=Bos taurus OX=9913 GN=KRT5 PE=2 SV=1 | KRT5 | 63 kDa | 0.67% |
| 1.3 | Keratin 3 OS=Bos taurus OX=9913 GN=KRT3 PE=1 SV=1 | KRT3 | 64 kDa | 0.64% |
| 1.4 | IF rod domain-containing protein OS=Bos taurus OX=9913 GN=KRT6A PE=3 SV=1 | KRT6A | 61 kDa | 0.47% |
| 1.5 | KRT4 protein OS=Bos taurus OX=9913 GN=KRT4 PE=2 SV=1 | KRT4 | 58 kDa | 0.27% |
| 1.6 | Keratin, type II cytoskeletal 75 OS=Bos taurus OX=9913 GN=KRT75 PE=2 SV=1 | KRT75 | 59 kDa | 0.27% |
| 1.7 | Keratin 77 OS=Bos taurus OX=9913 GN=KRT77 PE=1 SV=1 | KRT77 | 63 kDa | 0.24% |
| 1.8 | Keratin, type II cytoskeletal 79 OS=Bos taurus OX=9913 GN=KRT79 PE=3 SV=1 | KRT79 | 57 kDa | 0.17% |
| 2 | Cluster of Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 (P13645) | KRT10 | 59 kDa | 2.60% |
| 2.1 | Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 | KRT10 | 59 kDa | 2.60% |
| 2.2 | Keratin, type I cytoskeletal 15 OS=Ovis aries OX=9940 GN=KRT15 PE=2 SV=1 | KRT15 | 49 kDa | 0.44% |
| 3 | Keratin, type II cytoskeletal 1 OS=Homo sapiens OX=9606 GN=KRT1 PE=1 SV=6 | KRT1 | 66 kDa | 2.30% |
| 4 | Carbonic anhydrase 3 OS=Bos taurus OX=9913 GN=CA3 PE=2 SV=3 | CA3 | 29 kDa | 1.40% |
| 5 | Cluster of Keratin, type I cytoskeletal 14 OS=Bos taurus OX=9913 GN=KRT14 PE=1 SV=3 (F1MC11) | KRT14 | 56 kDa | 1.20% |
| 5.1 | Keratin, type I cytoskeletal 14 OS=Bos taurus OX=9913 GN=KRT14 PE=1 SV=3 | KRT14 | 56 kDa | 1.10% |
| 5.2 | Keratin, type I cytoskeletal 17 OS=Bos taurus OX=9913 GN=KRT17 PE=3 SV=1 | KRT17 | 49 kDa | 0.71% |
| 5.3 | Keratin 42 OS=Bos taurus OX=9913 GN=KRT42 PE=1 SV=1 | KRT42 | 50 kDa | 0.54% |
| 6 | Keratin, type I cytoskeletal 9 OS=Homo sapiens OX=9606 GN=KRT9 PE=1 SV=3 | KRT9 | 62 kDa | 0.98% |
| 7 | Phosphoglucomutase-1 OS=Bos taurus OX=9913 GN=PGM1 PE=3 SV=1 | PGM1 | 62 kDa | 0.91% |
| 8 | Trypsin OS=Sus scrofa OX=9823 PE=1 SV=1 | 24 kDa | 0.81% | |
| 9 | Fructose-bisphosphate aldolase OS=Bos taurus OX=9913 GN=ALDOA PE=1 SV=1 | ALDOA | 39 kDa | 0.54% |
| 10 | Desmoplakin OS=Bos taurus OX=9913 GN=DSP PE=1 SV=2 | DSP | 332 kDa | 0.57% |
| 11 | Pyridoxal phosphate homeostasis protein OS=Bos taurus OX=9913 GN=PLPBP PE=2 SV=1 | PLPBP | 30 kDa | 0.44% |
| 12 | L-lactate dehydrogenase A chain OS=Bos taurus OX=9913 GN=LDHA PE=2 SV=2 | LDHA | 37 kDa | 0.44% |
| 13 | Glutathione S-transferase A4 OS=Bos taurus OX=9913 GN=GSTA4 PE=2 SV=1 | GSTA4 | 26 kDa | 0.34% |
| 14 | Flavin reductase (NADPH) OS=Bos taurus OX=9913 GN=BLVRB PE=1 SV=1 | BLVRB | 21 kDa | 0.30% |
| 15 | Junction plakoglobin OS=Bos taurus OX=9913 GN=JUP PE=1 SV=1 | JUP | 81 kDa | 0.34% |
| 16 | NAD(P)H dehydrogenase, quinone 1 OS=Bos taurus OX=9913 GN=NQO1 PE=2 SV=1 | NQO1 | 31 kDa | 0.24% |
| 17 | Desmoglein-1 OS=Bos taurus OX=9913 GN=DSG1 PE=4 SV=2 | DSG1 | 112 kDa | 0.24% |
| 18 | Hemoglobin subunit beta OS=Bos taurus OX=9913 GN=HBB PE=1 SV=1 | HBB | 16 kDa | 0.13% |
| 19 | GTP:AMP phosphotransferase AK3, mitochondrial OS=Bos taurus OX=9913 GN=AK3 PE=1 SV=3 | AK3 | 26 kDa | 0.17% |
| 20 | Carboxymethylenebutenolidase homolog OS=Bos taurus OX=9913 GN=CMBL PE=4 SV=1 | CMBL | 28 kDa | 0.17% |
| 21 | Carbonic anhydrase OS=Bos taurus OX=9913 GN=CA2 PE=1 SV=3 | CA2 | 29 kDa | 0.13% |
| 22 | Uncharacterized protein OS=Bos taurus OX=9913 GN=GSTM2 PE=4 SV=2 | GSTM2 | 26 kDa | 0.20% |
| 23 | Pyruvate kinase OS=Bos taurus OX=9913 GN=PKM PE=1 SV=1 | PKM | 58 kDa | 0.13% |
| 24 | Keratin 24 OS=Bos taurus OX=9913 GN=KRT24 PE=3 SV=3 | KRT24 | 55 kDa | 0.17% |
| 25 | Annexin A2 OS=Bos taurus OX=9913 GN=ANXA2 PE=1 SV=2 | ANXA2 | 39 kDa | 0.10% |
| 26 | Peroxiredoxin-1 OS=Bos taurus OX=9913 GN=PRDX1 PE=2 SV=1 | PRDX1 | 22 kDa | 0.10% |
| 27 | Hydroxyacylglutathione hydrolase, mitochondrial OS=Bos taurus OX=9913 GN=HAGH PE=1 SV=1 | HAGH | 61 kDa | 0.10% |
| 28 | Triosephosphate isomerase OS=Bos taurus OX=9913 GN=TPI1 PE=3 SV=1 | TPI1 | 31 kDa | 0.10% |
| 29 | Glutathione S-transferase OS=Bos taurus OX=9913 GN=GSTM3 PE=1 SV=1 | GSTM3 | 28 kDa | 0.13% |
| 30 | Glycerol-3-phosphate dehydrogenase [NAD(+)] OS=Bos taurus OX=9913 GN=GPD1 PE=3 SV=1 | GPD1 | 49 kDa | 0.10% |
| 31 | Uncharacterized protein OS=Bos taurus OX=9913 GN=LOC100847119 PE=1 SV=2 | LOC100847119 | 25 kDa | 0.07% |
| 32 | Serum albumin OS=Bos taurus OX=9913 GN=ALB PE=4 SV=1 | ALB | 69 kDa | 0.20% |
| 33 | Glyceraldehyde-3-phosphate dehydrogenase OS=Bos taurus OX=9913 GN=GAPDH PE=1 SV=1 | GAPDH | 41 kDa | 0.10% |
| 34 | Plakophilin-1 OS=Bos taurus OX=9913 GN=PKP1 PE=4 SV=2 | PKP1 | 80 kDa | 0.07% |
| 35 | GLOBIN domain-containing protein OS=Bos taurus OX=9913 GN=HBA1 PE=3 SV=1 | HBA1 | 14 kDa | 0.07% |
| 36 | Peroxiredoxin-2 OS=Bos taurus OX=9913 GN=PRDX2 PE=2 SV=1 | PRDX2 | 22 kDa | 0.07% |
| 37 | Four and a half LIM domains 1 OS=Bos taurus OX=9913 GN=FHL1 PE=1 SV=2 | FHL1 | 36 kDa | 0.07% |
| 38 | LIM domain binding 3 OS=Bos taurus OX=9913 GN=LDB3 PE=4 SV=1 | LDB3 | 66 kDa | 0.13% |
| 39 | GTP-binding protein SAR1b OS=Bos taurus OX=9913 GN=SAR1B PE=2 SV=1 | SAR1B | 22 kDa | 0.07% |
| 40 | Creatine kinase M-type OS=Bos taurus OX=9913 GN=CKM PE=1 SV=2 | CKM | 43 kDa | 0.07% |
| 41 | Hydroxyacyl-CoA dehydrogenase OS=Bos taurus OX=9913 GN=HADH PE=4 SV=1 | HADH | 34 kDa | 0.07% |
| 42 | Actin, cytoplasmic 1 OS=Bos taurus OX=9913 GN=ACTB PE=1 SV=1 | ACTB | 42 kDa | 0.10% |
| 43 | Aconitate hydratase, mitochondrial OS=Bos taurus OX=9913 GN=ACO2 PE=1 SV=1 | ACO2 | 85 kDa | 0.07% |
| 44 | Filamin A interacting protein 1 like OS=Bos taurus OX=9913 GN=FILIP1L PE=4 SV=3 | FILIP1L | 135 kDa | 0.07% |
| 45 | Keratin, type II cytoskeletal 80 OS=Bos taurus OX=9913 GN=KRT80 PE=3 SV=1 | KRT80 | 49 kDa | 0.07% |
Table S3 (previous 8): Scaffold file of mass spectrum of 42-kD excised protein band.
| # | Identified Proteins (31) | Alternate ID | Molecular Weight | Percentage of Total Spectrum |
| 1 | Creatine kinase B-type OS=Bos taurus OX=9913 GN=CKB PE=1 SV=1 | CKB | 43 kDa | 4.20% |
| 2 | Serum albumin OS=Bos taurus OX=9913 GN=ALB PE=4 SV=1 | ALB | 69 kDa | 0.45% |
| 3 | Cluster of Keratin, type II cytoskeletal 1 OS=Homo sapiens OX=9606 GN=KRT1 PE=1 SV=6 (P04264) | KRT1 | 66 kDa | 0.56% |
| 3.1 | Keratin, type II cytoskeletal 1 OS=Homo sapiens OX=9606 GN=KRT1 PE=1 SV=6 | KRT1 | 66 kDa | 0.49% |
| 3.2 | Keratin 1 OS=Bos taurus OX=9913 GN=KRT1 PE=1 SV=2 | KRT1 | 63 kDa | 0.15% |
| 4 | Cluster of Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 (P13645) | KRT10 | 59 kDa | 0.37% |
| 4.1 | Keratin, type I cytoskeletal 10 OS=Homo sapiens OX=9606 GN=KRT10 PE=1 SV=6 | KRT10 | 59 kDa | 0.37% |
| 4.2 | Keratin, type I cytoskeletal 14 OS=Bos taurus OX=9913 GN=KRT14 PE=1 SV=1 | KRT14 | 50 kDa | 0.15% |
| 4.3 | Keratin, type I cytoskeletal 25 OS=Bos taurus OX=9913 GN=KRT25 PE=2 SV=1 | KRT25 | 49 kDa | 0.08% |
| 5 | Keratin, type I cytoskeletal 9 OS=Homo sapiens OX=9606 GN=KRT9 PE=1 SV=3 | KRT9 | 62 kDa | 0.37% |
| 6 | Acetyl-CoA acetyltransferase, mitochondrial OS=Bos taurus OX=9913 GN=ACAT1 PE=2 SV=1 | ACAT1 | 45 kDa | 0.37% |
| 7 | Aspartate aminotransferase, cytoplasmic OS=Bos taurus OX=9913 GN=GOT1 PE=1 SV=3 | GOT1 | 46 kDa | 0.30% |
| 8 | Fumarylacetoacetase OS=Bos taurus OX=9913 GN=FAH PE=3 SV=1 | FAH | 45 kDa | 0.22% |
| 9.1 | Keratin, type II cytoskeletal 2 epidermal OS=Homo sapiens OX=9606 GN=KRT2 PE=1 SV=2 | KRT2 | 65 kDa | 0.22% |
| 9.2 | IF rod domain-containing protein OS=Bos taurus OX=9913 GN=KRT6A PE=3 SV=1 | KRT6A | 61 kDa | 0.08% |
| 9.3 | Keratin 3 OS=Bos taurus OX=9913 GN=KRT3 PE=1 SV=1 | KRT3 | 64 kDa | 0.04% |
| 10 | Fructose-bisphosphate aldolase OS=Bos taurus OX=9913 GN=ALDOA PE=1 SV=1 | ALDOA | 39 kDa | 0.15% |
| 11 | Trypsin OS=Sus scrofa OX=9823 PE=1 SV=1 | 24 kDa | 0.19% | |
| 12 | Vinculin OS=Bos taurus OX=9913 GN=VCL PE=1 SV=1 | VCL | 111 kDa | 0.11% |
| 13 | Cluster of SERPIN domain-containing protein OS=Bos taurus OX=9913 PE=3 SV=2 (F1MMS7) | 45 kDa | 0.19% | |
| 13.1 | SERPIN domain-containing protein OS=Bos taurus OX=9913 PE=3 SV=2 | 45 kDa | 0.15% | |
| 13.2 | SERPIN domain-containing protein OS=Bos taurus OX=9913 GN=LOC511106 PE=3 SV=3 | LOC511106 | 44 kDa | 0.15% |
| 14 | Creatine kinase M-type OS=Bos taurus OX=9913 GN=CKM PE=1 SV=2 | CKM | 43 kDa | 0.15% |
| 15 | Cluster of Aldo-keto reductase family 1 member B1 OS=Bos taurus OX=9913 GN=AKR1B1 PE=1 SV=2 (P16116) | AKR1B1 | 36 kDa | 0.11% |
| 15.1 | Aldo-keto reductase family 1 member B1 OS=Bos taurus OX=9913 GN=AKR1B1 PE=1 SV=2 | AKR1B1 | 36 kDa | 0.11% |
| 15.2 | Aldo_ket_red domain-containing protein OS=Bos taurus OX=9913 PE=4 SV=1 | 24 kDa | 0.08% | |
| 16 | Acyl-CoA dehydrogenase long chain OS=Bos taurus OX=9913 GN=ACADL PE=2 SV=1 | ACADL | 48 kDa | 0.15% |
| 17 | Aldo-keto reductase family 1 member A1 OS=Bos taurus OX=9913 GN=AKR1A1 PE=2 SV=1 | AKR1A1 | 37 kDa | 0.08% |
| 18 | 3-ketoacyl-CoA thiolase, mitochondrial OS=Bos taurus OX=9913 GN=ACAA2 PE=1 SV=1 | ACAA2 | 42 kDa | 0.08% |
| 19 | Creatine kinase U-type, mitochondrial OS=Bos taurus OX=9913 GN=CKMT1 PE=2 SV=1 | CKMT1 | 47 kDa | 0.08% |
| 20 | Citrate synthase, mitochondrial OS=Bos taurus OX=9913 GN=CS PE=1 SV=1 | CS | 52 kDa | 0.11% |
| 21 | Cathepsin D OS=Bos taurus OX=9913 GN=CTSD PE=3 SV=1 | CTSD | 42 kDa | 0.08% |
| 22 | Ribonuclease/angiogenin inhibitor 1 OS=Bos taurus OX=9913 GN=RNH1 PE=1 SV=1 | RNH1 | 49 kDa | 0.08% |