Abstract
The first protein must produce at random processes. However, it is difficult to replicate correct protein without any control. The activities of control must be different from activities to be control. It is known that organisms replicate proteins via ribosomes by using genetic information. The mechanism that had replicated proteins naturally is a bridge between living organism and nonliving organism. We can make assumptions about structure of proto-ribosome on the base of its functions. That is, a proto-ribosome would have sandwiched L-type mRNA and D-type tRNA in part of a phospholipid bilayer. This structure can be used to estimate the initial processes of replicated proteins and the initial formation of aminoacyl-tRNA synthetases.
Keywords
Enzyme world, Ribosome, DNA, mRNA, tRNA, Aminoacyl-tRNA synthetase
Introduction
The first life had formed in non-extreme environment on the Earth because the first cell with gene system must have naturally formed. However, many of traditional studies of the origin life have been carried out on extremophiles [1], because most of professional researchers must acquire budgets, they have been studied by the acquired budget. The organisms had been born, then evolved by adaptations in its environment. By the results of evolution, extremophiles can live only in extreme environment [2]. On the other hand, there are numerous descriptions on molecular biology [3]. Especially ongoing progress in the structural biology is giving a physico-chemical basis that explains facets about tRNA [4]. Replication of protein is controlled via the informational media of double helix of DNA discovered by Watson and Click [5]. Karasawa reported that the replication processes of DNA are possible to reveal based on the structure of DNA [6,7]. The initial process of protein replication can be revealed based on the structure of the proto-ribosome which includes mRNA and tRNA in a part of a phospholipid bilayer. Here, the mRNA is left-handed (L-type) chirality, while the tRNA is right-handed (D-type). Although these two strands with different chirality enter a plathome of processing of central part of the double-layer, those strands never coalesce. The double-layered helical structures are indispensable in the ribosome-translation machinery.
Preparations
Formation of mRNA, tRNA and DNA
Organic molecules such as hydrocarbons were accumulated on the surface of water, and macroscopic boundary conditions formed a membrane [6]. When amino acid molecules adhered to the membrane, those molecules were formed molecular structures possessing with the function of enzymes. The first organization of life had formed in the world of enzymes. Current protein is replicated in a ribosome via short-lived mRNA and tRNA. Those mRNA and tRNA are produced from a replicated DNA [7]. Since RNA is a tool to deal with genetic information, the first life should be discussed in the real world of enzymes instead of the informational world of RNA.
Matching Processes between mRNA and tRNA
When amino acids adhere a membrane, conformation of a part of membrane is modified. The changed conformation of the membrane includes information on the amino acid sequence of a protein. Even though a proto-DNA is formed by simplifications from the membrane by exclusion of the protein, it possesses information of size and segmentation on the amino acid. Such information is used for the first matching processes between template of mRNA and matching objects of tRNA. Subsequent evolutions, pairing relationships for the matching was established by complementary base pairs of codons and anticodons. So, intermittently fixing of a segment of mRNA for a specified amino acid in a protein, each tRNA is shifted along the mRNA in order to looking for the partner of hydrogen bonds.
Leading Strand and Lagging Strand of DNA
Since chirality of mRNA is L-type but tRNA is D-type, two kinds of RNA do not merge. It is known that the chirality of biomolecule in the Earth, amino acids are L-type, and sugars are D-type. So, the membrane adhered with protein and bases has L-type chirality, but D-type of helical structure will be formed due to antagonistic and organized movements of interconnected helix structures [7]. That is, alternate rotations around X axis through the center of tetrahedron units changes the shape of tetrahedral unit projected in the X-Y plane from square to trapezoidal. When a pair of atoms of tetrahedron located at the end of the long and short site vibrate up-and-down movements, inner and outer in the tetrahedron’s vertices vibrate opposite directions. Under an assumption of such antagonistic movements, leading strand and lagging strand of DNA are synthesized simultaneously at each segmentation of the amino acid. Since each amino acid has individual size, the segment of constituent of ribosome for an amino acid is the same. tRNA is formed by a single D-type of lagging strand with base due to chirality difference between amino acid and lagging strand. So, the leading strand replicates continuously, whereas the lagging strand replicates discontinuously forming short fragments. Since bases of two RNAs touch via hydrogen bonds, tRNA is possible to move independently from mRNA. Here, the complementary anticodon of tRNA is vertical flip symmetry of corresponding amino acid of mRNA.
Results
Formation of Proto-ribosome in a Phospholipid Bilayer
Phospholipid contains a chiral center at C2 position of a glyceryl moiety [7]. Twisted phospholipids laterally interlock, and the interlock induces systematic motions due to systematic thermal vibrations of atoms [8]. A double layer sandwiched between two layers of hydrophilic heads spontaneously forms. If one of the layers in part of the bilayer is rotated by 180°, the progress of the helix is changed from the output side to the input side at the center of the bilayer. This bilayer, despite having two chirality centers, provides a one-directional screw movement by the one-directional rotation. When only mRNA enters the bilayer, it passes through the bilayer. However, if tRNA enters from the other side of the mRNA, both strands come into conflict at the central portion owing to chirality [7]. mRNA and the series of tRNA’s are sandwiched between a protein and a series of amino acids with base pairs facing each other at the center. Figure 1 is an illustration of a structure of proto-ribosome and its constituents proposed in this paper.
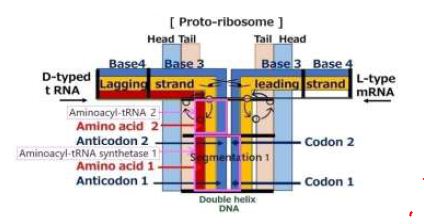
Figure 1: A structure of proto-ribosome and with its constituents
Prospect of the Protein Replication: Evolutions of Gene System on Chain Reactions
When a biological reaction is performed, a new reaction occurs due to change of the situation caused by the reaction. A chain reaction will continue to circulate if it forms a loop. A relationship of “from demand to the supply” will be included in those chain reactions. In various chain reactions, protein molecules that express repeated chain reactions will be formed and the enzymes will be formed. The chain reactions those support the survival of life will be incorporated into genes system in the form of long DNA. Then, Prokaryote have evolved to Eukaryote by formation of a nucleus of the cell in order to memorize very long DNA.
Discussions
Molecular Mechanisms Underlying Ribosome Dynamics
A step of protein replication is proceeded by amino acid unit at platform of a ribosome. The triplet base pairs in a DNA for each amino acid are formed via pattern matching on hydrogen bond between codon of mRNA and anticodon of tRNA. The complementally base pairs are adenine with thymine (A-T) and cytosine with guanine (C-G) for each base step. Incidentally, an aminoacyl-tRNA synthetase makes linkage between the triplet code and an amino acid by the direct attachment of an amino acid and corresponding tRNA [9]. Tamura, Schimmel reported about non-enzymatic aminoacylation of an RNA minihelix [10]. Karasawa proposes a functional model of aminoacyl-tRNA synthetase that comes from a cover around the functional model of aminoacyl-tRNA as shown in Figure 1. The linkage between amino acid and the triplet is carried out by an aminoacyl-tRNA synthetase. The proto-DNA forms a unique conformation when interacting with amino acids. The proto-DNA must possess information on amino acid. The enzyme binds with specific molecules, resulting in a conformational change, and carries out function of the catalyst. However, even the base sequence of tRNA has been revealed, understanding the molecular mechanisms underlying tRNA dynamics is yet challenging [11].
Conclusions
The author proposes that the first life should be discussed in the real world of enzymes instead of the informational world of RNA. Over the course of evolution, if a new mechanism is added alongside conventional mechanisms that functioning during life activities, the new mechanism must coexist with the conventional system. Eventually, the new system that successes to survive will remain, and unnecessary system will disappear. The research based on the current system is difficult to reveal the disappeared structures. The proto-ribosome was estimated based on the necessity that shifts tRNA reversely for mRNA and confirms the matched amino acid sequences. We can describe fundamental functions of ribosome by assuming such simple initial structure and its environments. The bottom-up approaches based on acceptable assumptions are useful to reveal initial processes of protein replications. However, there is a gap between the proposed model and current nucleotide sequence models in the molecular biology. It is known that the evolution of living organisms has influenced the Earth’s atmosphere and geology. It is another desire of the author that the proposed functional models will become bridges between living organisms and the field of geology of the Earth.
References
- Merino N, Heidi SA, Diana PB, Jayme FB, Michael LW, et al. (2019) Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context, Front Microbiol. [crossref]
- von Hegner I (2021) Extreme exoworlds and the extremophile paradox, arXiv.org | Cornell University Library, September.
- Watson JD, et al. (2004) Molecular biology of the Gene, 5th Ed. Benjamin Cummings.
- Nakanishi K, Nureki O (2005) Recent progress of structural biology of tRNA processing and modification. Mol Cells 19: 157-166. [crossref]
- Watson JD, Click FH (1953) Genetical Implications of the Structure of Deoxyribonucleic Acid, Nature 171: 964-967.
- Karasawa S (2023) Origin of life in the water of the Earth. Geology, Earth and Marine Science 5: 1-7.
- Karasawa S (2023) Initial processes on replication of DNA by interactions of helical structured molecules-Origin of life in the water of the Earth (Ⅱ). Geology, Earth and Marine Sciences 5: 1-7.
- MacKenzie LE, Stachelek P (2021) The twists and turns of chiral chemistry. Nature Chemistry 13: 521-522.
- Ibba M, Soll D (2000) Aminoacyl-tRNA synthesis. Annual Review of Biochemistry 69: 617-650.
- Tamura K, Schimmel P (2004) Non-enzymatic aminoacylation of an RNA minihelix with an aminoacyl phosphate oligonucleotide. Nucleic Acids Symposium Series 48: 269-270.
- Giege R, Frank J, Joern P, Peter S, Claude S, et al. (2012) Structure of transfer RNAs: Similarity and variability. WIREs RNA 3: 37-61. [crossref]