DOI: 10.31038/JMG.2020312
Abstract
Background: Loss of heterozygosity (LOH) at the human leukocyte antigen (HLA) region could lead to erroneous homozygous HLA typing results.
Materials and methods: We investigated HLA typing on peripheral blood samples derived from a patient with acute myeloid leukemia (AML) at diagnosis and remission by Luminex microbead assay. LOH was analyzed by short tandem repeat (STR) analysis at markers of long arm and short arm of chromosome 6 and single-nucleotide polymorphism (SNP) array analysis. For DNA mixing test to define the detection threshold for heterozygous HLA genotypes, we selected 6 samples of homozygous HLA typing and 6 samples of heterozygous HLA typing.
Results: At diagnosis (blasts 77% in peripheral blood), HLA typing revealed A*11, B*15:02/88(B75), C*08:01/08, DRB1*08, DQB1*06. Short tandem repeat (STR) analysis of peripheral blood revealed segmental uniparental disomy (UPD) of chromosome 6 (LOH of marker at 6p22, but no LOH at other markers of short arm and long arm of chromosome 6). Analysis of LOH by single-nucleotide polymorphism (SNP) array analysis demonstrated no copy number variations, but LOH on chromosome 6 [34.6Mb] and on chromosome 13 [92.8Mb]. At remission, HLA typing was A*11, A*24, B*15:02/112(B75), B*40:01/22N (B60), C*07:02/32N, C*08:01/08, DRB1*08, DRB1*14, DQB1*05, DQB1*06. DNA mixing experiments revealed that the minimum threshold for detecting HLA heterozygosity using Luminex technology is < 70% homozygous sample.
Conclusion: We describe an erroneous homozygoug HLA typing due to LOH resulting from segmental UPD. We should avoid sampling of blood with more than 70% blasts for HLA typing to reduce erroneous homozygous HLA typing results.
Keywords
human leukocyte antigen typing; loss of heterozygosity; uniparental disomy; microbead assay
Introduction
The human leukocyte antigen (HLA) gene complex resides on chromosome 6p21. HLA typing is used for HLA matching of donor–recipient pairs in hematopoietic stem cell transplantation (HSCT), searching HLA matched platelet components, identification of humoral responses to donor antigens in solid organ transplantation, and personalized risk assessment of HLA-associated autoimmune diseases and adverse drug reactions. Multiple mechanisms are responsible for HLA phenotypes alterations: alteration in steps in the biosynthetic pathway of HLA membrane expression, mutations in or loss of beta 2-microglobulin gene or HLA genes, and loss of heterozygosity (LOH) at the HLA region [1–3]. LOH at the HLA region could lead to loss of one HLA haplotype and erroneous homozygous HLA typing results [4]. LOH is defined as the loss of one parent’s contribution to the cell and can be caused by deletion, gene conversion, mitotic recombination, or loss of chromosome or uniparental disomy (UPD).
Copy-neutral LOH, also referred as to UPD [5], is thus called because no net change in the copy number occurs in the affected individual. UPD cannot be detected by conventional cytogenetics. UPD results when both copies of a chromosome pair originate from one parent. This might result in homozygosity [6]. In UPD, a person receives two copies of a chromosome (complex UPD), or part of a chromosome (segmental UPD), from one parent and no copies from the other parent due to errors in meiosis I or meiosis II [7].
Microsatellite analysis of STR and SNP array technology allows the identification and mapping of LOH in AML patients with normal karyotype. LOH can be identified in cancers by noting the presence of heterozygosity at a genetic locus in an organism’s germline DNA, and the absence of heterozygosity at that locus in the cancer cells. This is often done using polymorphic markers, such as analysis of short tandem repeats (STRs) and single-nucleotide polymorphism (SNP) array analysis. With genome-wide SNP-based array analysis providing both copy number and allele-specific information, it is possible to search the genome for subtle copy number alterations and regions with loss of heterozygosity [8].
Higher density SNP array can be used effectively to detect small regions of chromosomal changes and provide more information regarding the boundaries of loss regions. The Affymetrix 750K SNP array, an array of 750,000 markers for copy number analysis which consist of 550,000 unique non-polymorphic probes and approximately 200,000 SNPs, provides high-density SNP coverage for LOH detection, with greater than 99 percent accuracy.
Materials and methods
Samples and Institutional review board clearances
Venous blood samples were collected from a patient with acute myeloid leukemia (AML) at diagnosis and at complete remission, and healthy blood donors who signed informed consent. Three mL of blood was collected in sterile tubes containing EDTA. Buccal swab cells were obtained from the AML patient at complete remission. Karyotype analysis of the bone marrow of the AML patient did not show any chromosomal abnormality. The Institutional Review Board of Taipei Veterans General Hospital approved this study.
DNA extraction and analysis of PCR- STR
Genomic DNA was extracted from blood samples or buccal cell swabs with Puregene DNA isolation kit (Gentra System, Minneapolis, MN, USA) according to the method recommended by the manufacturer. In this study, six representative STR markers (D6S289, D6S276, D6S257, D6S434, D6S292, D6S281) covering the 6p/6q arms of chromosome 6 including the HLA region were selected for LOH study.
The polymerase chain reaction (PCR) amplification has been performed according to the method previous described by Ramal et al. [9] : 1.20 ml of DNA; 1.00 ml of Primer Mix (5 mM each primer); 1.50 ml of 10 × PCR reaction buffer including MgCl2 15 mM (Boehringer Mannheim, Mannheim, Germany); 1.50 ml of dNTPs mix (250 mM each dNTP); 0.12 ml of Taq DNA polymerase (5 U/ml) (Boehringer Mannheim) and distilled, deionized water to 15 ml of final volume.
The polymerase chain reaction (PCR) schedule was adjusted to GeneAmp® 9700 cycler (Applied Biosystems, Singapore) as follows: The initial denaturation at 95°C for 2 minutes (hot start); ten PCR cycles including 94ºC for 0.5 minutes, 55ºC for 0.5 minutes, 72ºC for 0.5 minutes; twenty PCR cycles including 89ºC for 0.5 minutes, 55ºC for 0.5 minutes, 72ºC for 0.5 minutes; and a final extension at 72°C for 10 min. Short tandem repeat (STR) data were analyzed using ABI PRISM® 310 Genetic Analyzer (Applied Biosystems Inc., Foster City, CA, USA).
LOH analysis by STR markers of chromosome 6
STR markers (D6S289, D6S276, D6S257) of short arm and STR markers (D6S434, D6S292, D6S281) of long arm were analyzed. Positions of STR markers are summarized in Table 1.
Table 1. Results of loss of heterozygosity detected by markers of short tandem repeats at chromosome 6
|
STR marker |
Chromosome location |
LOH calculation |
Results |
|
D6S289 |
6p22.3 |
0.91 |
NO LOH |
|
D6S276 |
6p22 |
0.67 |
LOH |
|
D6S257 |
6p12.1 |
Non-informative |
Homozygous |
|
D6S434 |
6q16.3 |
1.02 |
NO LOH |
|
D6S292 |
6q23.3 |
0.91 |
NO LOH |
|
D6S281 |
6q27 |
0.93 |
NO LOH |
In capillary electrophoresis, the ratio of the amount of PCR product for the two alleles is derived from the relative peak heights. It was assigned LOH when more than 25% of signal reduction of one allele was observed in the analyzed samples as compared to the control sample [9].
Ratio of allele height (RH) = peak height of the smaller allele/ peak height of the larger allele
Reduction of signal = 1 – (RH of analyzed sample/RH of control sample)
LOH analysis by SNP array analysis
We used SNP array analysis to evaluate LOH in a specific chromosomal region. Detection of LOH requires SNPs to be heterozygous (i.e., informative). SNP analysis was performed using Affymetrix CytoScan® Assay (CytoScan® 750K Array, Affymetrix Inc. Taiwan) was performed following the manufacturer’s protocols [10]. After performing a SNP array hybridization experiment, each slide is scanned, and the array probe signal intensities and SNP calls are subsequently analyzed. The signal intensities are analyzed to determine copy number estimate with the updated version 2.0 of the CNAG (Copy Number Analyzer for Affymetrix GeneChip mapping) software package [11]. Copy number variants were analyzed by array comparative genomic hybridization.
Luminex technology for HLA typing by PCR-SSO
The LABType SSO (One Lambda, Inc., Canoga Park, CA, USA) assays for HLA typing was performed according to the method previous described by Trajanoski et al. [12]. Target DNA is polymerase chain reaction (PCR) amplified using group-specific primers and then biotinylated which allows it to be detected using R-Phycoerythrin-conjugated Streptavidin. The PCR product is then denatured and allowed to hybridise to complementary DNA probes conjugated to fluorescently code microsperes. The Luminex Flow Analyser was used to detect the fluorescent intensity on each microsphere. The assignment of HLA alleles is based on the reaction pattern of the various beads compared to patterns with known HLA alleles.
DNA mixing experiments
Six sample (sample A) of homozygous HLA typing at a locus (HLA-A*02; B*38; C*07) and six sample (sample B) of heterozygous HLA typing at the same locus (A*02, A*11;B*15, B*38; C*07,C*08) were selected. The concentrations of these samples were adjusted to 20 ng/microliter. Sample A and sample B were serial mixed as the following ratio: 1:9, 2:8, 3:7 and 4:6, respectively. HLA typing was performed for these mixture samples individually. The assay threshold is the minimum allowable concentration of the sample B at which the heterozygous HLA typing can be determined.
Results
HLA typing results at diagnosis of AML and remission
At diagnosis of AML (blasts 77% in peripheral blood), HLA typing by PCR-SSOP revealed A*11, B*15:02(B75), C*08:01/08, DRB1*08, DQB1*06. At remission, HLA typing results were A*11, A*24, B*15:02 (B75), B*40:01/22N (B60), C*07:02/32N, C*08:01/08, DRB1*08, DRB1*14, DQB1*05,DQB1*06. HLA typing on cells obtained by a buccal swab at remission revealed the same HLA typing results.
Assignment of LOH by STR
Results of loss of heterozygosity detected by markers of short tandem repeats at chromosome 6 were shown in Table 1. A clear LOH is observed in alleles of marker D6S276 which located at 6p22.3–21.3 near HLA gene complex resides within chromosome 6p21. But no LOH was observed in other alleles of markers: 6S289 (6p22.3), D6S434 (6q16.3),D6S292(6q23.3), D6S281(6q27). So segmental UPD of chromosome 6 is suggested.
Assignment of LOH by SNP Array
Results of genome-wide SNP analysis were shown in Figurer 1. LOH were observed on chromosome 6 [34.6Mb] and on chromosome 13 [92.8Mb], but no copy number variation in all chromosomes was demonstrated (Fig. 1).
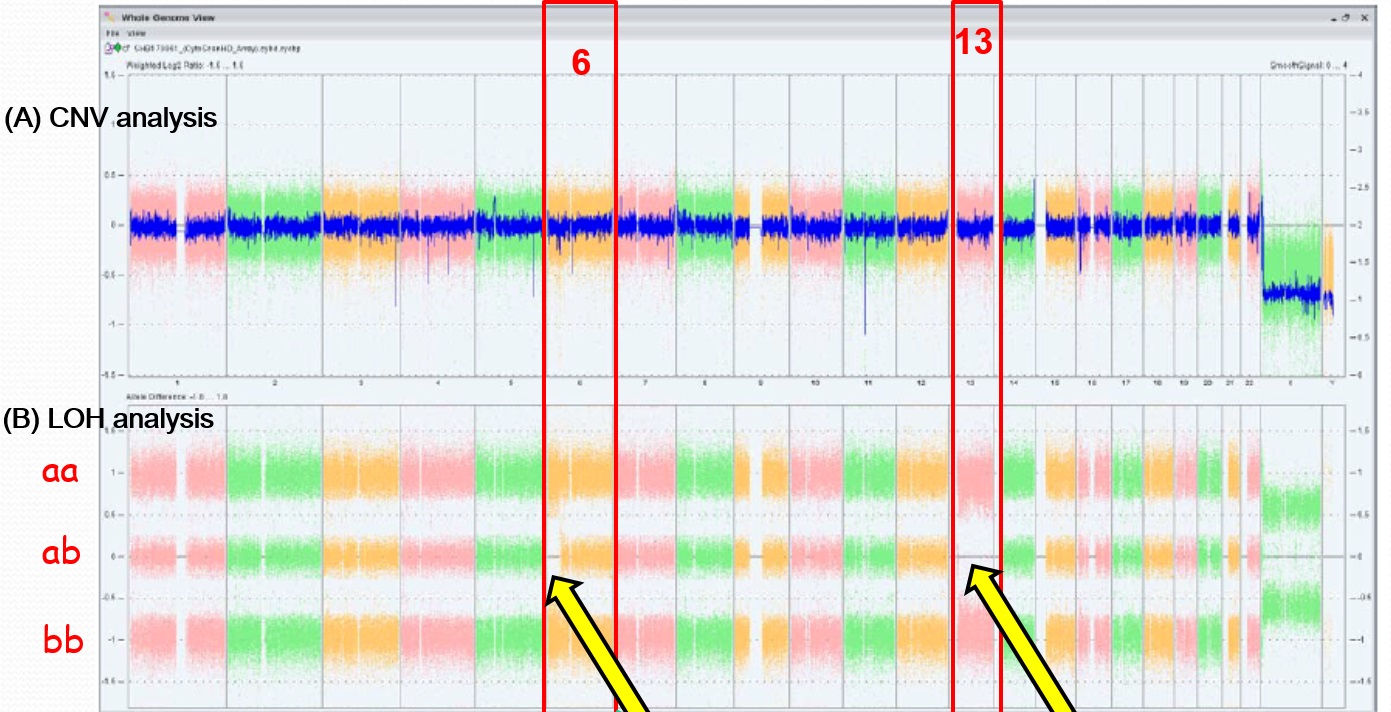
Figure 1. Results of single nucleotide polymorphism array analysis
X-axis represents number of chromosomes. (A) No copy number variation was observed in all chromosomes. (B) Loss of heterozygosity (LOH) was observed at short arm of chromosome 6 [34.6Mb] and chromosome 13 [92.8Mb]. CNV = copy number variation.
Detection threshold of HLA typing by Luminex
Using LABType SSO assays, the detection rate of heterozygous HLA typing for the mixture of a sample of homozygous HLA typing and a sample of heterozygous HLA typing at the same locus were summarized in Table 2. The threshold of LABType SSO assays for detecting heterozygosity was more than 30% of the samples with heterozygous HLA typing.
Table 2. Detection rate of heterozygous HLA genotype by DNA mixing experiments (n=6)
|
Heterozygous HLA type |
Ratio of sample B to sample A |
|||
|
1:9 |
2:8 |
3:7 |
4:6 |
|
|
HLA-A*11 |
0 |
83.3 |
100 |
100 |
|
HLA-B*15 |
16.7 |
83.3 |
100 |
100 |
|
HLA-C*08 |
0 |
50 |
100 |
100 |
Sample A: homozygous HLA typing of HLA-A*02; B*38; C*07;
Sample B: heterozygous HLA typing of HLA-A*02, A*11; B*15, B*38; C*07, C*08.
Discussion
We report false homozygous HLA genotyping in a patient with AML at diagnosis when the blast cell was 77% in blood sample. Microsatellite markers have shown segmental UPD. Copy number variant analysis by array comparative genomic hybridization did not reveal copy number variations in chromosome 6 and thus confirmed that the HLA homozygosity was due to partial UPD (Fig. 1). We repeated HLA genotyping at complete remission, and correct heterozygous HLA typing was obtained. In this case LOH was present at diagnosis, suggesting that clones with acquired UPD could occur spontaneously in the developing leukemia.
Systematic application of whole genome scanning technologies with SNP arrays has demonstrated that LOH without changes in copy number frequently occur in many types of cancer [13]. Bullinger et al. performed high-resolution SNP analyses in 157 adult cases of CN-AML, and regions of acquired UPDs were identified in 12% of cases and in the most frequently affected chromosomes, 6p, 11p and 13q [14]. Genome-wide analysis of SNPs in AMLs has revealed that 18.8 % of AML patients exhibited large regions of homozygosity due to partial UPD, and the homozygosity was found to be restricted to the leukemic clone [15]. The role of UPD may be underestimated. Raghavan et al. found an increased frequency of UPD (41%) at relapsed AML [16]. After transplantation of haploidentical hematopoietic stem cells the CNN-LOH in 6p provides a common mechanism of leukemic relapse after HLA haploidentical stem cell transplantations, in which leukemic cells can escape the immunologic surveillance of the engrafted donor T cells through the loss of the mismatched HLA haplotype [17, 18].
By DNA mixing test, we found a low proportion (< 30%) of homozygous cells cannot be detected with Luminex technology. LOH can involve the entire HLA region, but in some cases it may be partial with the involvement of only one or a few HLA loc. Dubois et al. studied 6 AML patients with HLA mistyping, and found that complete HLA-A, B, C homozygosity happened in patients with more than 80% blast cells except in one case of acute myeloid leukemia in which LOH was observed only at locus A with 27% of blast cells but pronounced monocytosis [4]. It is unusual to observe LOH in patients without blast cells. Partial remission may be associated with peripheral blood leucocytes affected by chromosomal abnormalities without morphological malignancy [19].
Conclusions
In conclusion, we reported erroneous HLA typing resulted from segmental UPD of chromosome 6 in an AML patient at diagnosis with blasts 77% in peripheral blood sample. Blood sample with more than 70% blast cells should not be used for HLA typing to reduce erroneous HLA typing results by Luminex microbead assay.
Funding
This study was supported by Taipei Veterans General Hospital and Taiwan Clinical Oncology Research Foundation.
Authorship contributions
J.S. Lin and L.H. Lee wrote the manuscript. J.S. Lin, L.H. Lee, H.M. Liu, and T.J. Chiou designed the study. Y.J. Chen coordinated the study. L.H. Lee and H.M. Liu enrolled the subjects and performed the experiment. J.S. Lin and L.H. Lee analyzed the data and performed the statistics. All authors reviewed and approved the final version of the manuscript.
References
- Garrido F, Ruiz-Cabello F, Cabrera T, Perez-Villar JJ, Lopez-Botet M et al. (1997) Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today 18: 89–95. [crossref]
- Koopman LA, Mulder A, Corver WE, Anholts JD, Giphart MJ, et al. (1998) HLA class I phenotype and genotype alterations in cervical carcinomas and derivative cell lines. Tissue Antigens 51: 623–636. [crossref]
- Browning M, Dunnion D (1997) HLA and cancer: implications for cancer immunotherapy and vaccination. Eur J Immunogenet 24: 293–312. [crossref]
- Dubois V, Sloan-Bena F, Cesbron A, Hepkema BG, Gagne K, Gimelli S (2012) Pretransplant HLA mistyping in diagnostic samples of acute myeloid leukemia patients due to acquired uniparental disomy. Leukemia 26: 2079–2085. [crossref]
- Thomas LaFramboise T (2009) Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res 37: 4181–4193. [crossref]
- Tuna M, Knuutila S, Mills GB (2009) Uniparental disomy in cancer. Trends Mol Med 15: 120–128. [crossref]
- Kotzot D (2001) Complex and segmental uniparental disomy (UPD): review and lessons from rare chromosomal complements. J Med Genet 38: 497–507. [crossref]
- Komura D, Shen F, Ishikawa S, Fitch KR, Chen W et al. (2006) Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res 16: 157515–84. [crossref]
- Ramal LM, Feenstra M, van der Zwan AW, Collado A, Lopez-Nevot MA et al. (2000) Criteria to define HLA haplotype loss in human solid tumors. Tissue Antigens 55: 443–448 [crossref]
- McMullan DJ, Bonin M, Hehir-Kwa JY, de Vries BB, Dufke A et al. (2009) Molecular karyotyping of patients with unexplained mental retardation by SNP arrays: a multicenter study. Hum Mutat 30: 1082–1092. [crossref]
- Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L (2005) A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res 65: 6071–6079. [crossref]
- Trajanoski D, Fidler SJ (2012) HLA typing using bead-based methods. Methods Mol Biol 882: 47–65. [crossref]
- Makishima H, Maciejewski JP (2011) Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res 17: 3913–23. [crossref]
- Bullinger L, Kronke J, Schon C, Radtke I, Urlbauer K et al. (2010) Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 24: 438–449. [crossref]
- Raghavan M, Lillington DM, Skoulakis S, Debernardi S, Chaplin T (2005) Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res 65: 375–378. [crossref]
- Raghavan M, Smith LL, Lillington DM, Chaplin T, Kakkas I (2008) Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood 112: 814–821. [crossref]
- Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C (2009) Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 361: 478–488. [crossref]
- Villalobos IB, Takahashi Y, Akatsuka Y, Muramatsu H, Nishio N et al. (2010) Relapse of leukemia with loss of mismatched HLA resulting from uniparental disomy after haploidentical hematopoietic stem cell transplantation. Blood 115: 3158–3161. [crossref]
- Bontadini A, Iannelli S, Fruet F, Capelli S, Masetti R, Dubois V, et al. (2015) Erroneous HLA typing as a result of acquired uniparental disomy in a patient with acute lymphoblastic leukaemia in peripheral blood complete remission. Blood Transfus 13:678–681. [crossref]