DOI: 10.31038/JMG.2019214
Abstract
The post-translational acetylation of the histone components of chromatin mediates numerous DNA-templated events, including transcriptional activation, DNA repair, and genomic replication. The conserved SAGA (Spt-Ada-Gcn5 Acetyltranferase) and SLIK (SAGA-Like) Histone Acetyltransferase (HAT) complexes are required for transcriptional activation of a subset of yeast genes and contain multiple subunits including the histone fold-containing TBP- Associated Factors (TAFs): 6, 9, 10, and 12. These TAFs are also components of the TFIID complex and are consequently involved in most RNA polymerase II-transcription in yeast. Here we identify a novel conserved region of TAF12, termed ReNu, outside of its histone fold, which is required for SAGA and SLIK-directed nucleosomal acetylation. We demonstrate that this region is not required for chromatin association, but show that this region plays an important role for histone H3 acetylation at specific SAGA and SLIK-regulated promoters. Our data suggests that the ReNu region of TAF12 regulates Gcn5 acetylation of specific substrates within the SAGA super-family of HAT complexes.
Introduction
The packaging of the eukaryotic genome into chromatin is generally believed to be refractive to DNA-templated processes such as transcriptional activation, DNA repair, and replication. However, chromatin-modifying complexes regulate these processes through histone modifications such as acetylation, methylation, ubiquitination, and phosphorylation [1]. Histone acetylation correlates with transcriptional capacity and is catalyzed by histone acetyltransferase (HAT) activities such as the yeast Spt-Ada-Gcn5-Acetyltransferase (SAGA) coactivator complex [2]. Both yeast and human SAGA complexes contain the HAT subunit Gcn5 (or the related PCAF protein), the Tra1, Ada, Spt, and a subset of TAF proteins [3–8]. The histone fold containing TAFs 6, 9, 10, and 12 are components of the SAGA, SAGA-like (SLIK/SALSA), and TFIID complexes in yeast, a feature conserved through evolution [9]. TFIID and SAGA are broadly required for transcriptional activation [10,11], however specific targeting of the TAF12 subunit of these complexes has recently been found to lead to a selective transcriptional response that interferes with acute myeloid leukemia [12]. Prior work had shown that TAF12 can bind to the activation regions of the Gal4, GCN4, VP16 and HNF4A transcription factors and that these interactions make differential contributions to transcriptional activation [13–18].
The observation that TAF12 is required for SAGA HAT activity on nucleosomal substrates is complicated by the fact that a taf12 temperature sensitive mutation in yeast compromises SAGA structural integrity and size with the observed loss of at least the SAGA Spt3 subunit [4]. Here we examine the direct role of TAF12 in SAGA and SLIK-mediated HAT activity on specific substrates by assaying complexes bearing various mutations in TAF12. Furthermore, we investigate the in vivo contributions of an evolutionarily conserved TAF12 region on nucleosome acetylation at specific SAGA and SLIK regulated promoters.
Materials and Methods
Yeast Strains and Point Mutation Generation
SAGA and SLIK were prepared from yeast strain YSB493 (MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12-259::LEU2 {pRS314-TAF12}, YSB556(MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12- 259::LEU2 {pZM279-taf12 322-539}), YSB557 (MATa, ura3-52, leu2::PET56, trp1-1, his3- 200, ade2, taf12-259::LEU2 {pZM280-taf12 374-539}) [19] and YJR21-9 (taf12-9) [20].
TAF12 ReNu region point mutations were generated using the Quick Change Protocol (Stratagene) where pRS314-TAF12 served as a TAF12 template. Point mutation incorporated TAF12 plasmids were shuffled into yeast strain YSB452 (MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12-259::LEU2 {pJA73-TAF12-URA3}) to generate strains YMT1 (MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12-259::LEU2 {pRS314-taf12 N336A}), YMT2 (MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12-259::LEU2 {pRS314-taf12 L344A}) and YMT3 (MATa, ura3-52, leu2::PET56, trp1-1, his3-200, ade2, taf12-259::LEU2 {pRS314-taf12 S342A}).
Purification of Mutant SAGA and SLIK Complexes
SAGA and SLIK complexes were partially purified by a scheme adapted from our earlier work [4]. In brief, 4 liters of yeast were grown to mid-log phase at 30ºC. Yeast whole cell extracts were prepared by glass bead disruption [4]. Extracts were bound batch wise with 15ml of Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen). The resin was washed in a column with 20mM imidazole followed by elution with 300mM imidazole. The Ni2+-NTA agarose eluate was directly loaded onto a MonoQ HR 5/5 column (GE Healthcare). Bound proteins were eluted with a 25ml linear gradient from 100 to 500 mM NaCl. Peak SAGA fractions were pooled and concentrated down to 0.7ml with a Centriprep-30 concentrator (Millipore). Samples were then loaded on a Superose 6 H/R 10/30 column (GE Healthcare) equilibrated with 350mM NaCl. Peak fractions were used in subsequent assays.
Western Blotting and HAT Assays
Proteins from column fractionations were separated by 12% SDS-PAGE, transferred to nitrocellulose, and processed for immunoblotting using the antisera described. HAT assays were performed in the presence of 2 mg HeLa nucleosomes or core histones, 0.25 µCi of [3H]acetyl coenzyme A, and equivalent amounts of purified SAGA complexes, as described [4].
Chromatin Immunoprecipitation Assays
ChIP assays were performed largely as described [21]. Yeast strains YSB493 and YSB557 were grown in Synthetic Complete (SC) medium containing 2% dextrose to an OD 600 of 1.0. The yeast were reseeded in SC dextrose-medium, SC medium containing 2% galactose, or SC medium containing 2% KAc and then grown for an additional 4 hr. Antiserum specific for histone H3 acetylated at lysine 9 (Millipore) was used for ChIP. Input and precipitated DNA was PCR amplified for regions upstream of the CIT2, GAL10, RPL9A, and RPS5, as described in Pray-Grant MG et al [7]. Real-time PCR experiments were performed with three biological replicates and conducted with a SYBR Green/Taq polymerase reaction mix (Bio-Rad) in a MyIQ real-time PCR detection system (Bio-Rad). Data were collected and analyzed with thermal cycler software (Bio-Rad).
Results and Discussion
The Novel TAF12 ReNu Region Regulates SAGA and SLIK Nucleosomal Acetylation
Primary sequence analysis of yeast TAF12 reveals that the protein consists of three regions: an amino-terminal glutamine rich domain, followed by a novel-conserved region upstream of the TAF12 carboxy-terminal histone fold domain (Figure 1a). We examined the structural and functional roles of these domains by assaying SAGA complexes containing full length TAF12 (WT), a TAF12 mutant protein deleted of amino acids 1-322 spanning the non-conserved glutamine rich domain (QΔ,), and a TAF12 mutant deleted of both the glutamine rich domain and an additional 52 amino acids (QΔ + ReNuΔ), hereafter named ReNuΔ. Western analysis using antisera against multiple SAGA subunits reveal that the structural integrity of the partially purified SAGA complex was intact in both of the TAF12 truncation protein strains, and interestingly, Spt3 was identified as a component of SAGA in all complexes (Figure1b). This could be because the two histone folds of Spt3 may be free to interact with the remaining histone fold of TAF12 in either complex [22]. Ada2, Ada3, and Gcn5 also associate with the TAF12- truncated SAGA complexes (Figure 1b) and are important for SAGA HAT function [23–25]. We assayed WT, QΔ, and ReNuΔ SAGA for HAT activity on specific histone substrates. We found that deletion of the glutamine rich domain of TAF12 did not interfere with SAGA HAT activity on free histones or nucleosomes (Figure 1b). However, deletion of both the TAF12 glutamine rich domain and the additional 52 C-terminal amino acids abolished nucleosomal acetylation, while there was no effect on SAGA HAT activity on free histone substrates (Figure1b). This result suggests that within the SAGA complex, this novel TAF12 region regulates Gcn5 substrate specificity, and we have consequently named it the ReNu (Required for Nucleosomal acetylation) region. Consistent with this, invariant amino acids within the ReNu region contribute to SAGA nucleosomal acetylation (Figure 4b).
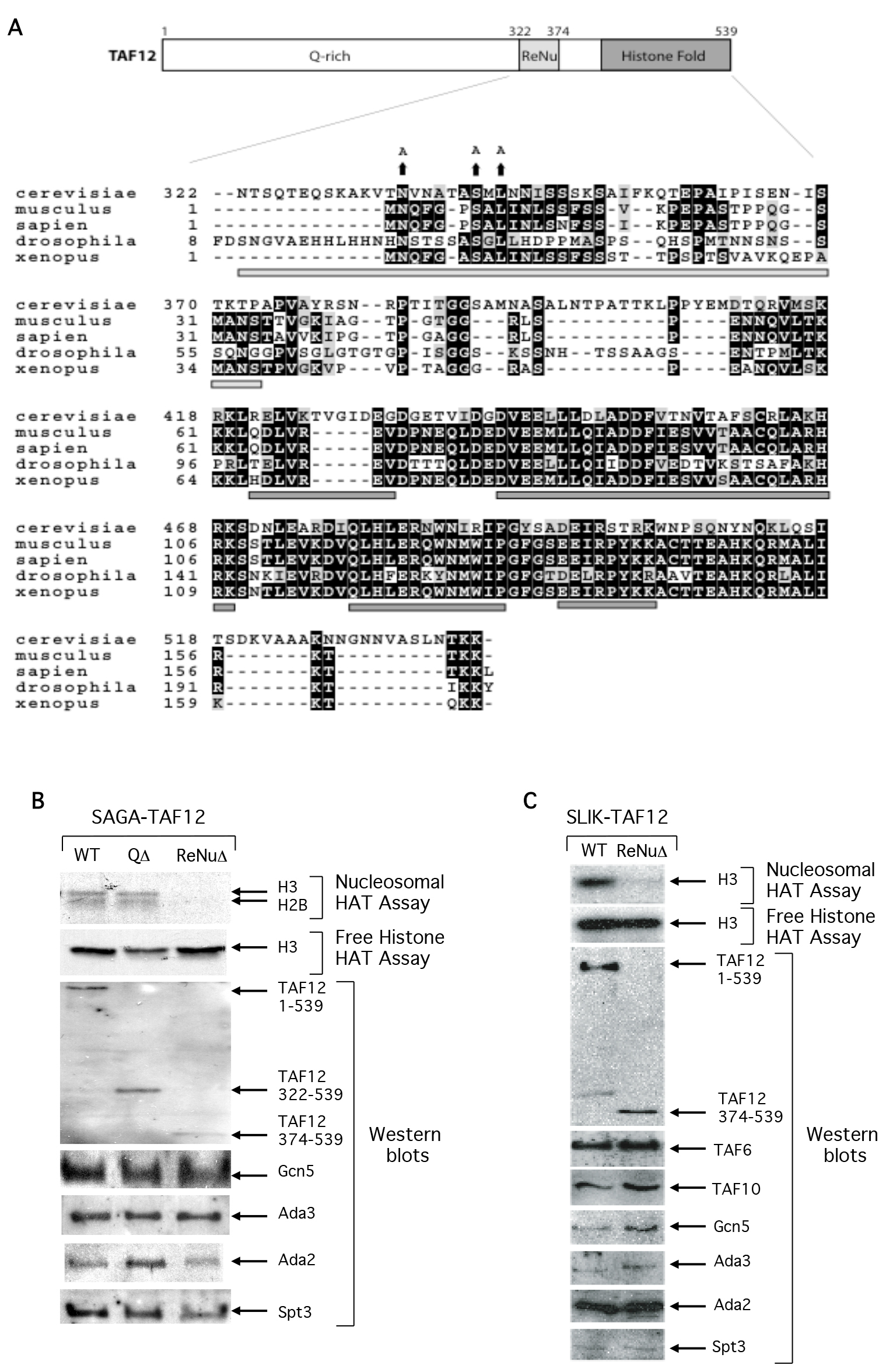
Figure 1. The TAF12 ReNu domain is required for SAGA-dependent nucleosome acetylation.
(A) Primary sequences of the Saccharomyces cerevisiae, Mus musculus, Homo sapien, Drosophila melanogaster, and Xenopus laevis TAF12 homologs were aligned in ClustalW and graphically represented with the BoxShade program. Conserved residues are shown in bold and similar residues are shaded. The regions encompassing the minimal ReNu region and four histone-fold α-helices, but excluding the yeast non-conserved glutamine rich region, are represented by bars underneath the alignment in light and dark grey respectively. The changed sequence of invariant amino acids in the TAF12 yeast mutants is shown above the corresponding wild-type residues. (B) HAT assays and Western blot analysis of partially purified SAGA complexes. SAGA was prepared from the yeast strains YSB493, expressing full length TAF12 (WT), YSB556, yeast deleted of the TAF12 glutamine rich N-terminus (QΔ), and YSB557, yeast deleted of the TAF12 glutamine rich and ReNu regions (ReNuΔ). HAT assays were performed with free histone or nucleosome substrates and Western analysis was performed with the indicated antisera. (C). HAT assays and Western blot analysis of partially purified SLIK complexes. SLIK was prepared from the yeast strains YSB493 and YSB557. HAT assays were performed with free histone or nucleosome substrates and Western analysis was performed with the indicated antisera.
The related SLIK complex also contains both Gcn5 and TAF12 [7,26]. Therefore, we examined the contribution of the ReNu region in SLIK-mediated nucleosomal acetylation. We found that the structural integrity of partially purified SLIK was intact in both WT and ReNuΔ complexes and that Spt3 associates with ReNuΔ SLIK (Figure 1c). Similar to ReNuΔ SAGA, ReNuΔ SLIK was found to acetylate free histones at a level comparable to WT levels, but was unable to acetylate nucleosomes (Figure 1c).
These data establish a nucleosome acetylation regulatory role for the TAF12 ReNu region within the SAGA and SLIK HAT complexes and help explain the acquired ability of Gcn5 to acetylate nucleosomal histones when present in native complexes [7,8,27]. This may be achieved by directly interacting with the core catalytic components of the complex, Gcn5, Ada2 or Ada3 [23,24] or through histone tail presentation in the context of the nucleosome. Notably, Ada3 is a key component in regulating Gcn5’s nucleosomal acetyltransferase activity, but is still insufficient to drive native SAGA dependent nucleosomal acetylation in the absence of functional TAF12.
Yeast Strains Lacking the TAF12 ReNu Region Exhibit a Modest gcn5 Phenotype
We examined the role of the ReNu region in genetic assays, hypothesizing that yeast lacking the TAF12 ReNu region may display a gcn5 phenotype, due to the loss of SAGA nucleosomal acetylation. We assayed wild-type and ReNu deleted yeast strains for growth defects on media containing low nitrogen, potassium acetate as a carbon source, and caffeine [7,8]. Serial dilutions of a ReNu wild-type strain, a ReNu-deleted yeast strain and a gcn5 strain were compared. We found that in these media, yeast bearing a deletion of the ReNu region exhibit a modest growth defect, not as severe as that of a gcn5 strain (Figure 2). All strains showed equivalent growth on the rich media YPD (Figure 2). Presumably, the observed intermediate gcn5 phenotype may be because TAF12 is not present in all yeast Gcn5 containing complexes [28]. Therefore, the observed modest phenotype might be a result of compensation of SAGA/SLIK acetyltransferase activity by other Gcn5 containing complexes, such as ADA and HAT-A2.
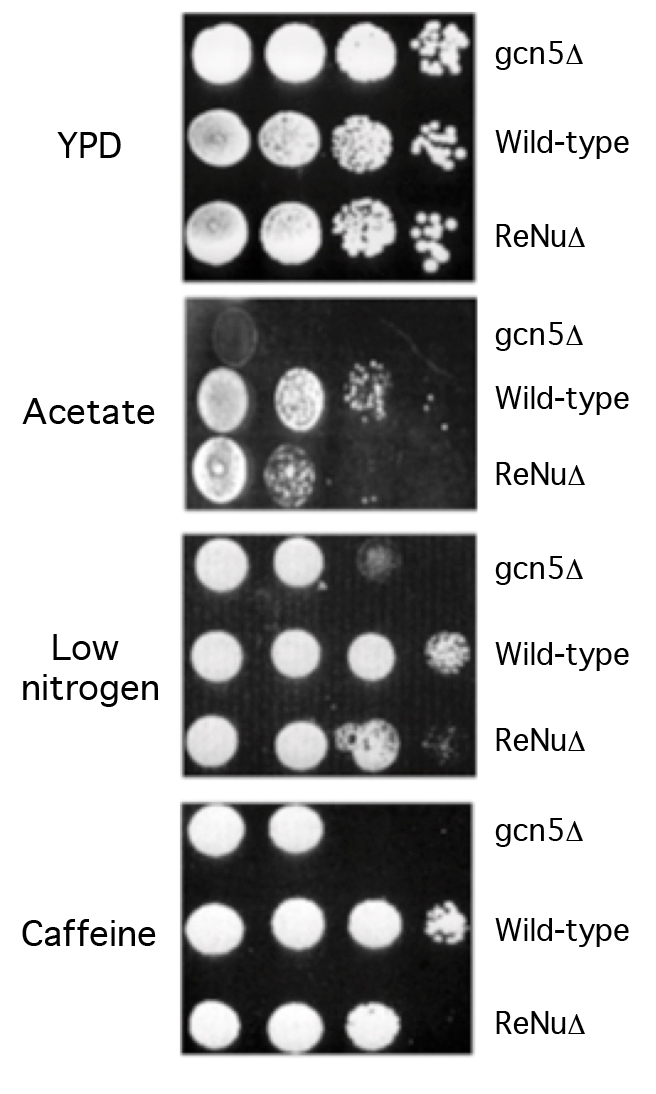
Figure 2.The TAF12 ReNu domain mediates Gcn5 function in vivo.
Wild-type, gcn5Δ and TAF12 ReNuΔ yeast strains were assayed for their ability to grow on Yeast extract-Peptone-Dextrose (YPD) media, synthetic complete media with 2% Potassium Acetate as a carbon source (Acetate), synthetic complete media supplemented with 12mM caffeine, and synthetic complete media lacking ammonium sulphate (low nitrogen). Shown are agar plates with ten-fold serial dilutions of each yeast strain, from left to right.
The TAF12 ReNu Region is not Required for SAGA Substrate Associations
The observed loss of nucleosomal acetylation in ReNuΔ SAGA and SLIK could be due to a loss of chromatin association. We examined this possibility by binding ReNu WT or ReNuΔ SAGA complexes to SONs (short, 3–6, oligonucleosomes) and then fractionating these reactions by size exclusion chromatography. Fractions were processed for western analysis and immunoblotted with histone H4 antisera in order to detect the presence of SONs in any given fraction. We hypothesized that unbound SONs should fractionate at later fractions than those SONs bound to SAGA because the SAGA-SON interactions would shift SONs on a size exclusion column to an earlier fraction, corresponding to a higher molecular mass. As predicted, unbound SONs fractionated with an apparent mass of 0.6–1.0 MDa (fraction 22–24). Both ReNu WT and ReNuΔ SAGA shifted SONs towards the 2MDa column exclusion volume, fraction 18/19, which correlates to a mass at or above that predicted for SAGA, 1.8 MDa (Figure 3A). This data suggests that the ReNuΔ SAGA complexes retain the ability to associate with nucleosomes and furthermore, this association is salt stable in 350mM NaCl buffer.
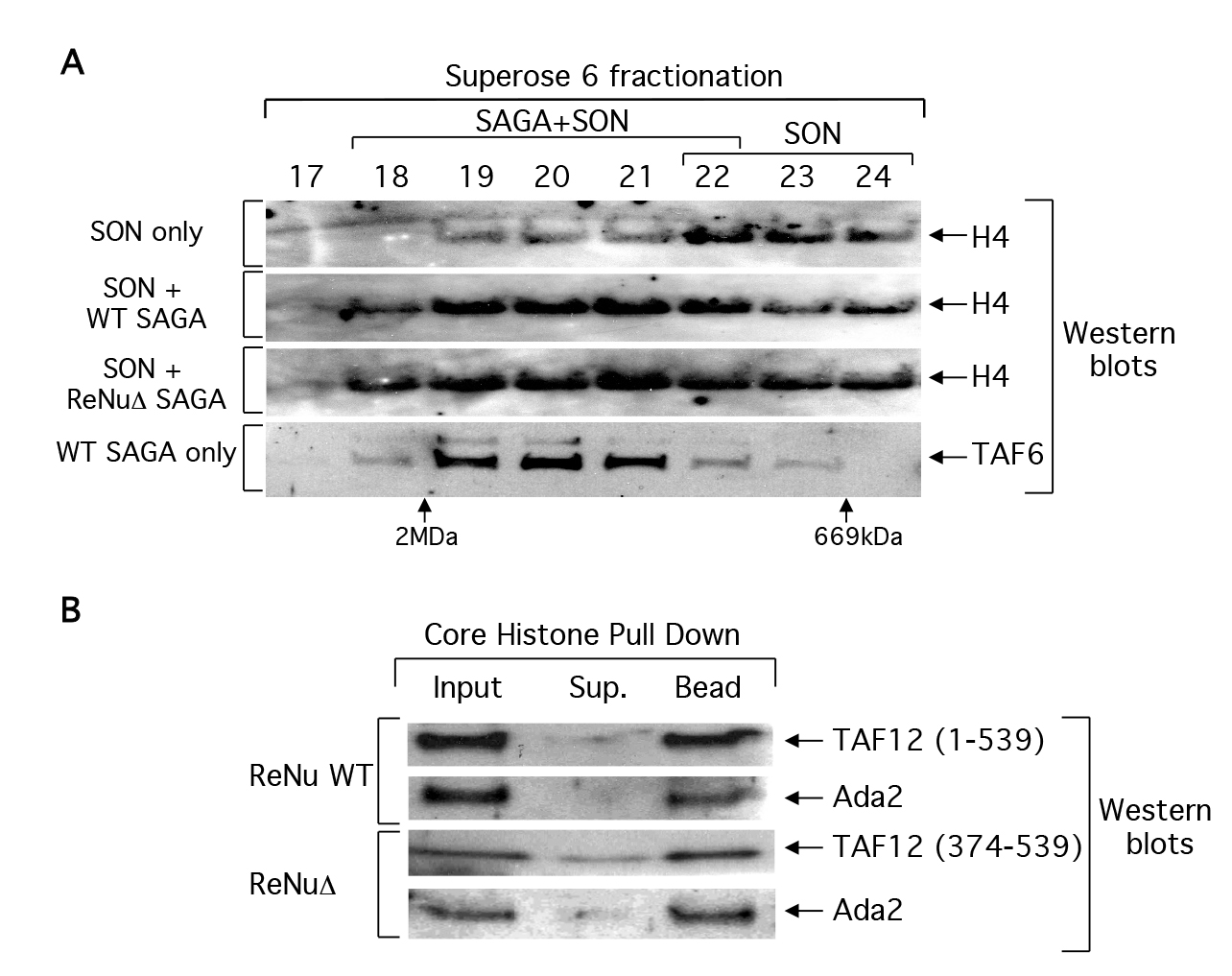
Figure 3. ReNu deleted SAGA interacts with nucleosomes and core histones.
(A) Partially purified wild-type (WT) and ReNuΔ SAGA complexes were incubated with short oligonucleosomes (SON) and then fractionated over a Superose 6 size exclusion column. Fractions were processed for western blotting and the presence of SON were detected with histone H4 antiserum, and the elution of SAGA was monitored with TAF6 antiserum. Elution of the dextran blue (2MDa) and thyroglobulin (669kDa) molecular mass markers from the same column is indicated. (B) WT SAGA and ReNuΔ SAGA were bound to immobilized core histones. Inputs, supernatants (sup), and bound fractions (bead) from the reactions were analyzed by western blotting with the indicated antisera to SAGA components
We further assayed whether or not immobilized core histones could pulldown SAGA in standard binding assays. We bound WT SAGA and ReNuΔ SAGA to core histone agarose resin and analyzed the bound fractions with antisera against SAGA subunits. Similar to our SAGA- nucleosome interaction data, we observed that ReNuΔ SAGA can interact efficiently with core histones (Figure 3b) or double stranded DNA (data not shown). Taken together, our data demonstrate that ReNuΔ SAGA retains nucleosomal and core histone substrate interactions and that the loss of nucleosomal acetylation by ReNuΔ SAGA is likely not due to a loss in substrate association.
Invariant Amino Acids within the ReNu Region Contribute to SAGA Nucleosomal Acetylation
SAGA complexes were purified from yeast bearing point mutations in invariant amino acids: N336A, S342A, and L344A (Figures 4a and 4b), as described [7,27]. Western analysis indicates that similar to ReNuΔ SAGA, point mutant SAGA complexes retained their structural integrity and predicted native size of a 1.8 Mda (Figure 4a). Also similar to ReNuΔ SAGA, the three point mutants had little effect on SAGA’s free histone acetylation (Figure 4b). However, each significantly reduced nucleosomal acetylation by SAGA (Figure 4b) albeit not as completely as ReNuΔ. Additionally, ReNu point mutant yeast were serially diluted on low nitrogen and found to give an intermediate phenotype between our ReNu WT and ReNuΔ yeast (Figure 4c). Together, these findings indicate that each of the three invariant residues within the ReNu region play an important role in SAGA-mediated nucleosomal acetylation.
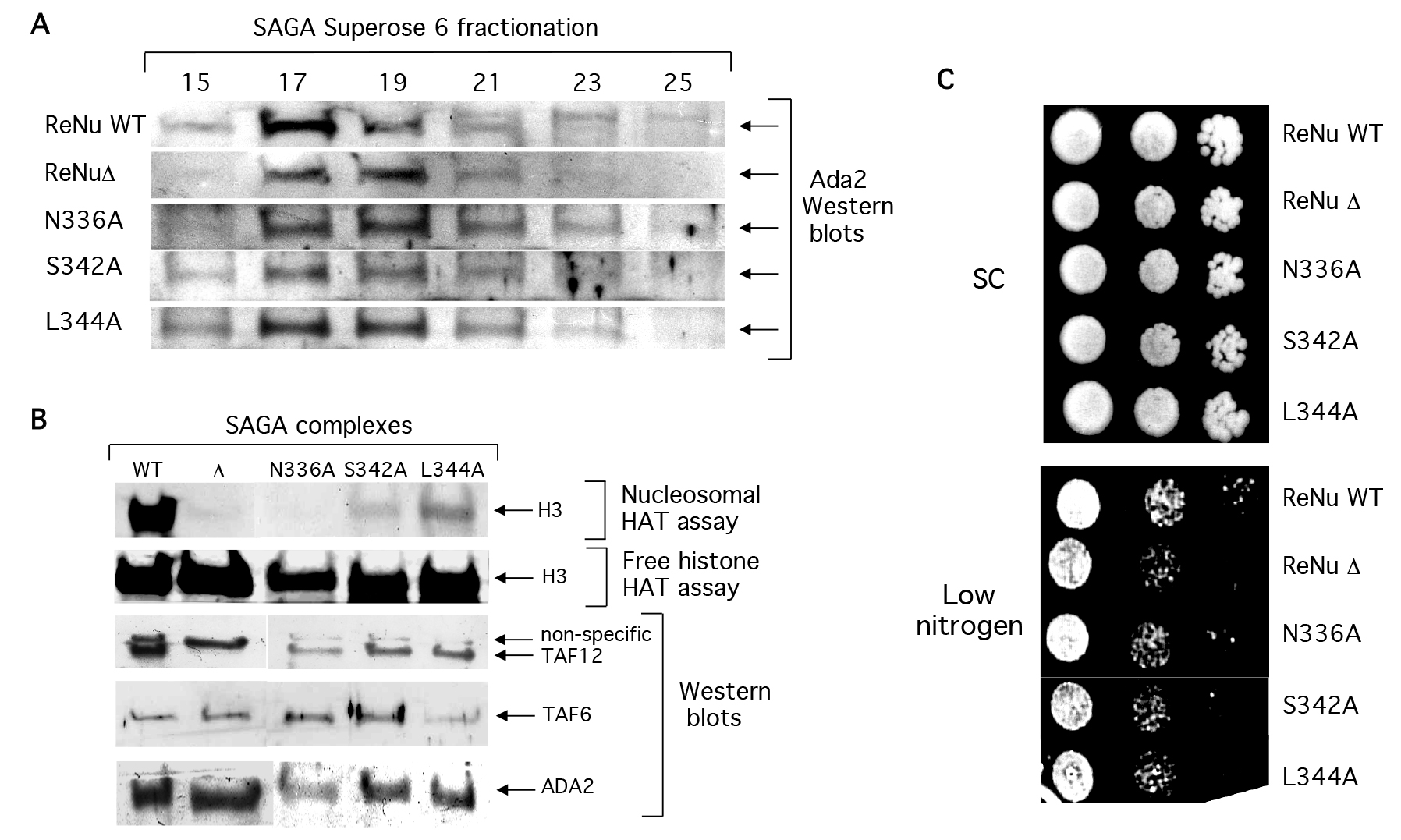
Figure 4. ReNu region point mutations compromise SAGA catalytic function, but not structural integrity.
(A) Western blot analysis of partially purified WT ReNu, ReNuΔ, or ReNu point mutant SAGA complexes. SAGA was prepared from the yeast strains YSB493, YSB557, YMT1 (taf12 N336A), YMT2 (taf12 L344A), and YMT3 (taf12 S342A). Yeast extracts were sequentially purified by nickel-NTA, anion exchange, and size exclusion chromatography. SAGA fractionated in superose 6 fractions 17–19 as determined by Ada2 immunoblotting, in an approximately 1.8 MDa complex. (B) SAGA complexes were similalrly partially purified from the yeast strains and peak fractions were incubated in HAT assays with nucleosome and free histone substrates. Western blotting from the same fractions was performed with the indicated antisera. (C) ReNu region point mutant yeast N336A, S342A, and L3444A were serially diluted on low nitrogen and found to give an intermediate phenotype relative to ReNu WT and ReNuΔ yeast.
The TAF12 ReNu Region is Essential for Promoter Acetylation
To further investigate the in vivo role of the TAF12 ReNu region in nucleosomal acetylation and to validate our in vitro observations, we examined specific promoter acetylation in wild type ReNu and ReNu region deleted yeast strains. Therefore, we conducted Chromatin Immunoprecipitation (ChIP) assays to assess the acetylation status of SAGA and SLIK-regulated promoters GAL1 and CIT2 [7,29–32]. Under inducing conditions, the GAL1-10 UAS is occupied by the SAGA and SLIK complexes [7,32–35], SAGA/SLIK is required for GAL1 and GAL10 mRNA production [36,37], and CIT2 expression is regulated by SLIK [38]. ChIP assays were performed using antisera against H3K9Ac (acetylated lysine 9 of histone H3) because this modification is implicated as being dependent on Gcn5 [39,40]. Upon activation, WT ReNu yeast show increased H3 K9Ac signal at both the GAL1-10 and CIT2 UAS. However, we found a striking absence of H3K9Ac signal at GAL1-10 and CIT2 UAS in ReNuΔ yeast (Figure 5a). These findings are in agreement with our in vitro data showing a loss of nucleosomal acetylation in ReNuΔ SAGA and SLIK. Interestingly, H3K9Ac ChIPs for the RPS5 and RPL9A promoters, both described as TFIID-regulated genes [36], showed no loss of acetylation in the ReNuΔ yeast (Figure 5a). These results suggest that the TAF12 ReNu region is required for H3K9Ac at the SAGA and SLIK- regulated genes GAL1-10 and CIT2, but not at the TFIID-regulated genes RPS5 and RPL9A. To quantify the changes in acetylation observed in TAF12 mutant strains we performed real-time PCR for the GAL-10 UAS under inducing conditions. We performed ChIP in gcn5Δ, or TAF12 mutant strains for H3K9Ac relative to wild-type strains. These experiments revealed a reduction in acetylation to approximately 55% in gcn5Δ (p=0.003) and 63% in ReNuΔ strains (p=0.01), while deletion of the Q-rich region caused no reduction in acetylation (Figure 5b).
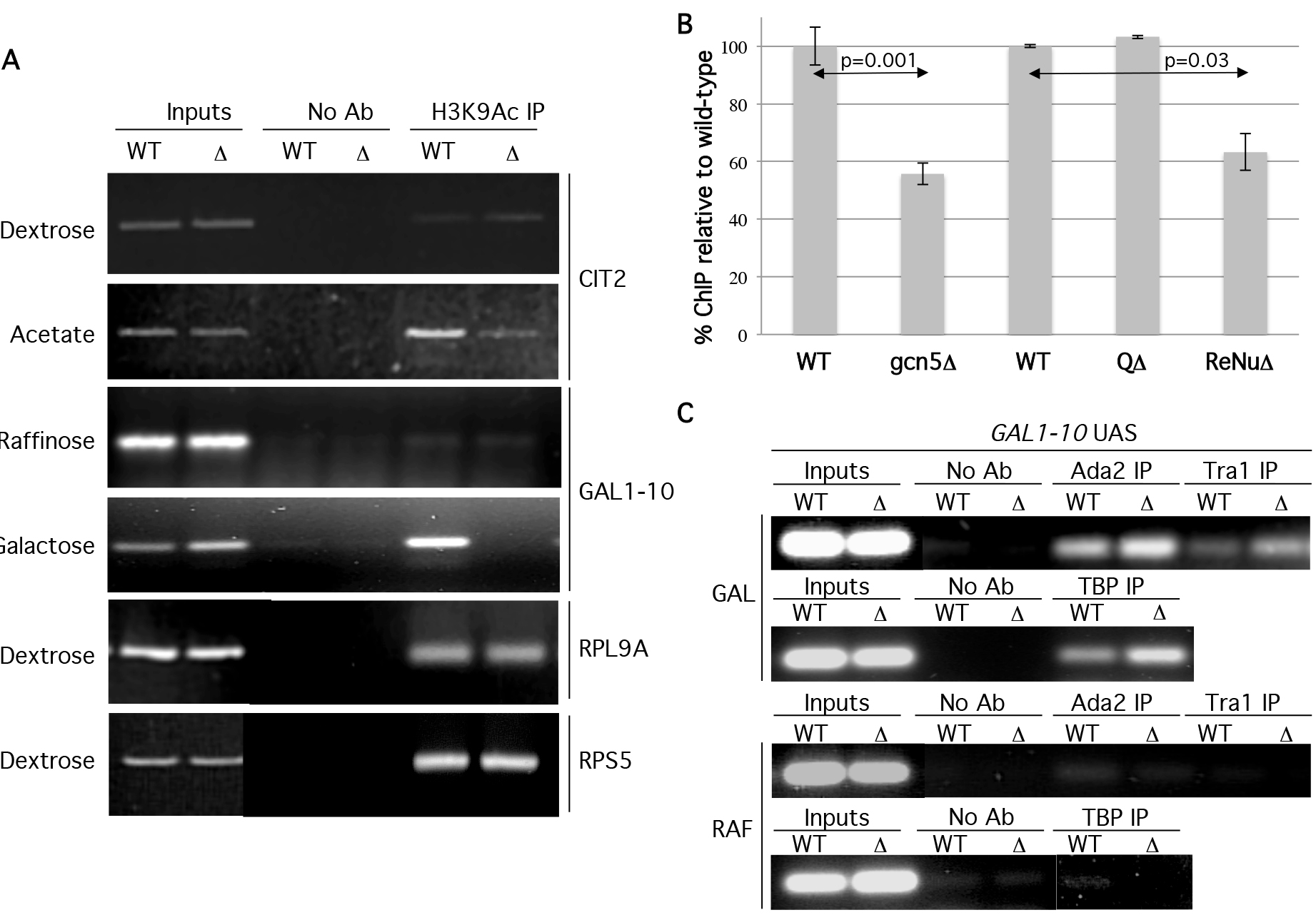
Figure 5. The ReNu region is required for H3K9Ac at SAGA and SLIK regulated UAS.
(A) ChIP assays were performed from wild type (WT) or ReNu-deleted (Δ) yeast under noninducing dextrose (Dex) or raffinose (Raff) and inducing potassium acetate (KAc) and galactose (Gal) conditions, respectively, with antisera against H3K9Ac. Precipitated DNA was amplified by PCR using primers for the gene loci indicated. A control IP was performed without antibody (no Ab). (B) Down regulation of H3K9 acetylation at the GAL-10 UAS in gcn5Δ and ReNuΔ strains under inducing (Gal) conditions was quantified using Real-time PCR. Error bars represent standard errors and p-values were derived from two-tailed Student t tests. (C). ChIP assays were performed under noninducing (RAF) or inducing (GAL) conditions with antisera against Tra1, Ada2, and TBP. Precipitated DNA was amplified by PCR using primers for the GAL-10 UAS.
The loss of H3K9Ac at the GAL1-10 and CIT2 UAS in ReNuΔ yeast could be caused by an inability of SAGA and SLIK to interact with the promoter-chromatin environment. SAGA and SLIK components are recruited to the GAL1-10 UAS during activation [7,32–35]. We tested this possibility by using antisera against SAGA and SLIK components Ada2 and Tra1 in ChIP assays at the GAL1-10 UAS. As expected, we found Ada2 and Tra1 were recruited to the GAL-10 UAS in ReNu WT yeast under inducing conidtions. When we examined ReNuΔ yeast, we not only found that we could ChIP ReNuΔ SAGA/SLIK to the GAL1-10 UAS, but that there was a modest enrichment of Ada2 and Tra1 to the GAL1-10 locus (Figure 5c). These data, taken together with our findings that ReNuΔ SAGA interacts with nucleosomes, (Figure 3a) suggest that the observed loss of H3K9Ac at GAL1 is not caused by an inability of SAGA or SLIK to interact with the GAL1 UAS in vivo. Furthermore, our studies agree with findings [34] which demonstrate that SAGA is recruited to the GAL1 UAS in the absence of the Gcn5, which is equivalent to the loss of SAGA-mediated nucleosomal acetylation found in ReNuΔ complexes.
SAGA possesses multiple activities besides its nucleosomal acetylation function such as TATA binding protein (TBP) recruitment via Spt3 and Spt8 [32,41–43], acidic activator association [32,33,44–47], deubiquitination of histone H2B [35,48], and methyl H3K4 binding [26,49]. We examined a SAGA function that is independent of its nucleosome HAT activity in order to determine whether or not ReNuΔ SAGA and SLIK were compromised in other complex functions. We examined SAGA and SLIK’s ability to recruit TBP to the GAL1 UAS [30,36,50,51] by ChIP using an antiserum against TBP in WT and ReNuΔ yeast. Our data reveal that both WT and ReNuΔ SAGA/SLIK recruit TBP to the GAL1 locus, and interestingly, TBP is modestly enriched at this locus in the ReNuΔ strain (Figure 5b). This could be explained through the enhanced GAL1 UAS residency of the SAGA and SLIK complexes also found in the ReNuΔ yeast. Furthermore, a similar result shows that a taf12 ts strain shows a slight enrichment of TBP at GAL1 and that TBP can still be recruited to the GAL1 UAS in gcn5Δ yeast, which lack Gcn5-mediated nucleosomal acetylation [36].
ReNuΔ yeast are capable of growing in galactose because the GAL1 gene is still transcribed in the absence of Gcn5 [42,50]. In addition, the loss of nucleosomal acetylation at the GAL1 and CIT2 UAS is likely not caused by compromised HAT complex recruitment via acidic activators because studies have shown that the acidic activator GAL4 can bind the GAL1 UAS in the absence of the SAGA subunits Spt3, Spt20, or Gcn5 [42]. This suggests that the acetylation function or structural integrity of SAGA is not required for GAL4 association with the GAL1 promoter. Also, Rtg3, the acidic activator required for CIT2 expression has been shown to interact with the CIT2 UAS independently of SAGA or SLIK [52] and Rtg3 protein levels are comparable between WT, gcn5Δ, or ada2Δ yeast strains [53].
The SAGA/SLIK component Spt3 recruits TBP [32,42,43] and TAF12 is required for Spt3 association with the SAGA complex [4]. Here we show that Spt3 is retained in ReNuΔ SAGA, but there is a loss of nucleosomal acetylation, indicating that the effects we observe are relevant to TAF12 rather than Spt3 function. The increased occupancy of mutant SAGA at target promoters seen in our ChIP assays may indicate that substrate acetylation is required for dissociation of the SAGA complex. Furthermore, TBP is also recruited at GAL1-10 at higher levels in ReNuΔ yeast. One possible explanation for this is that Mot1 may not be able to remove TBP from the GAL1-10 UAS in ReNuΔ yeast. It has been reported that Mot1 occupancy at the GAL1 promoter is greatly reduced in a gcn5 yeast strain [54].
Conclusion
Our results describe a novel regulatory function for a TAF protein, which is mediated by a conserved disordered region outside of the histone fold domain. The in vitro nucleosomal acetylation defects observed with ReNu-lacking SAGA complexes agree with the in vivo loss of acetylation at SAGA and SLIK-regulated promoters. Complexes bearing wild-type or truncated TAF12 are capable of interacting with DNA, histones and nucleosomes in in vitro binding studies, indicating that the ReNu region is not necessary for substrate interaction.
Together these data suggest that TAF12 mediates a regulatory mechanism for modulating Gcn5 HAT activity in SAGA and SLIK. This may be achieved by directly interacting with the core catalytic components of the complex, Gcn5, Ada2 or Ada3 [23,24] or through histone tail presentation in the context of the nucleosome. These findings may also implicate a putative mechanism by which HAT complexes are activated, which would likely be conserved in all other members of the SAGA HAT superfamily: SLIK, PCAF, STAGA, and TFTC. It is noteworthy that some of the conserved amino acids in the ReNu region have been described as sites for post-translational modification, including serines 342, 348 and 352 in S.cerevisiae and threonine 283 in S.pombe [55–58], which may serve to regulate ReNu function and correspondingly Gcn5 activity.
Data Availability
Data not included within this manuscript are available from the corresponding author upon request.
Footnotes
Abbreviations used are: SAGA, Spt-Ada-Gcn5-acetyltransferase; TAF, RNA polymerase II Specific TATA-binding protein associated factors; ReNu, TAF12 region required for nucleosomal acetylation; SLIK, SAGA-like histone acetyltransferase; HAT, histone acetyltranse complex; GCN5, general control non-derepressible; PCAF, p300/CBP-associated factor; ADA, alteration/deficiency in activation; SPT, suppressors of Ty transcription; TBP, TATA-binding protein; TFTC, TBP-free TAF containing; STAGA, Spt3-TAF31-Gcn5L acetyltransferase; SC, synthetic complete media; YPD, yeast extract-peptone-dextrose media, ChIP, chromatin immunoprecipitation assay; PCR, polymerase chain reaction; NTA, Ni2+-nitrilotriacetic acid
Acknowledgement
We thank Drs Joe Reese, Stephen Buratowski, Shelley Berger, and Jerry Workman for yeast strains and antisera, and Drs C. David Allis, David Auble, and the members of the Grant lab for technical advice and stimulating discussion. P.A.G. is supported by NIH grant R01 GM111911. This work was previously supported by NIH grant R01 DK58646 to P.A.G.
References
- Suganuma T, Workman JL (2011) Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80: 473–499. [crossref]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650.
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, et al. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell 94: 35–44. [crossref]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR., Workman JL (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45.
- Brand M, Yamamoto K, Staub A, Tora L (1999) Identification of TATA-binding protein- free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289.
- Martinez E, Kundu TK, Fu J, Roeder RG (1998) A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem 273: 23781–23785. [crossref]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, et al (2002) The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol 22: 8774–8786.
- Sterner DE, Belotserkovskaya R, Berger SL (2002) SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci 99: 11622–11627.
- Green MR (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci 25: 59–63.
- Baptista T, Grunberg S, Minoungou N, Koster MJ E, Timmers HTM, et al (2017) SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol Cell 2017.
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, et al. (2017) Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Mol Cell 68: 118–129. [crossref]
- Xu Y, Milazzo JP, Somerville TDD, Tarumoto Y, Huang YH, et al (2018) TFIID-SAGA Perturbation that Targets MYB and Suppresses Acute Myeloid Leukemia. Cancer cell 33: 13–28 e18
- Alpern D, Langer D, Ballester B, Le Gras S, Romier C, Mengus G, Davidson I (2014) TAF4, a subunit of transcription factor II D, directs promoter occupancy of nuclear receptor HNF4A during post-natal hepatocyte differentiation. eLife 3:03613.
- Fishburn J, Mohibullah N, Hahn S (2005) Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18, 369–378
- Hall DB, Struhl K (2002) The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem 277: 46043–46050.
- Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, et al. (2010) Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol 30: 2376–2390. [crossref]
- Klein J, Nolden M, Sanders SL, Kirchner J, Weil PA, Melcher K (2003) Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J Biol Chem 278: 6779–6786.
- Reeves WM, Hahn, S (2005) Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25: 9092–9102.
- Moqtaderi Z, Yale JD, Struhl K, Buratowski S (1996) Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci U S A 93: 14654–14658. [crossref]
- Reese JC, Green MR (2001) Genetic analysis of TAF68/61 reveals links to cell cycle regulators. Yeast 18: 1197–1205. [crossref]
- Kuo MH, Allis CD (1999) In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 19: 425–433.
- Birck C, Poch O, Romier C, Ruff M, Mengus G, et al (1998) Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94: 239–249.
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942.
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S (2002) Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem 277: 7989–7995. [crossref]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, et al. (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98. [crossref]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR., Grant PA (2005) Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, et al (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650.
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem 274: 5895–5900.
- Wu PY, Winston F (2002) Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol 22: 5367–5379.
- Deckert J, Struhl K (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol Cell Biol 21: 2726–2735.
- Bhaumik SR, Green MR (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 288: 1935–1945. [crossref]
- Larschan E, Winston F (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956. [crossref]
- Bhaumik SR, Green MR (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15: 1935–1945. [crossref]
- Bryant GO, Ptashne M (2003) Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell 11: 1301–1309. [crossref]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, Grant PA (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871.
- Bhaumik SR, Green MR (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22: 7365- 7371.
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR., Grant PA (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J.Biol.Chem. 279: 1867.
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, Grant, P. A. (2002) The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol.Cell.Biol. 22: 8774.
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY (1998) Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. Embo J 17: 3155–3167.
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem 274: 5895–5900.
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, et al (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701.
- Dudley AM, Rougeulle C, Winston F (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13: 2940–2945. [crossref]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F (1992) SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331.
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, et al. (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337. [crossref]
- Kuo MH, vom Baur E, Struhl K, Allis CD (2000) Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell 6: 1309- 1320.
- Allard S, Utley RT, Savard J, Clarke A, Grant P, et al (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. Embo J 18: 5108–5119.
- Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG (1999) Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell 4: 657–664.
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, et al. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663. [crossref]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, et al. (2011) Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842. [crossref]
- Topalidou I, Papamichos-Chronakis M, Thireos G (2003) Post-TATA binding protein recruitment clearance of Gcn5-dependent histone acetylation within promoter nucleosomes. Mol Cell Biol 23: 7809–7817.
- Li XY, Bhaumik SR, Green MR (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288: 1242–1244.
- Jia Y, Rothermel B, Thornton J, Butow RA (1997) A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 17: 1110–1117.
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, et al. (1999) A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell 4: 63–73. [crossref]
- Topalidou I, Papamichos-Chronakis M, Thireos G, Tzamarias D (2004) Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. Embo J 23: 1943–1948.
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396.
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686.
- Laboucarié T, Detilleux D1, Rodriguez-Mias RA2, Faux C1, Romeo Y, et al. (2017) TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability. EMBO Rep 18: 2197–2218. [crossref]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, et al (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nature methods 10: 676–682.