DOI: 10.31038/CST.2017252
Abstract
A normal cell has several processes that balance the process of self-renewal and differentiation. Multiple influences on gene expression orchestrate an orderly process resulting in normal tissue growth and function. Heritable changes in gene expression of a cell are called epigenetic changes. Epigenetics is a relatively new field in cancer studies and is increasingly being recognized to play a signification role in cancer formation and progression. Several epigenetic processes have been discovered ranging from DNA methylation to non-coding RNA. Epigenetic changes in a cancer cell are intriguing as they lend themselves to being targeted and provide avenues for cancer therapeutics.
Keywords
Epigenetics; cancer; chromatin; histone; noncoding RNA
Introduction
Epigenetics is the study of phenotypical changes occurring in a cell, with no change in the DNA sequence. The epigenetic creation of a new phenotype or state happens through specific mechanisms that affect gene expression in a stable, heritable manner. To further understand what the true meaning of epigenetics is and what it entails, a comparison to genetics is helpful. Genetics is the study of heredity and genes. Heredity is the process of passing on certain characteristics and traits through DNA from one generation to the next. In this case, phenotypes – or the traits and characteristics – are dependent upon the DNA sequence – or genotypes – an organism gets from the previous generation. Epigenetics is similar, in that it encompasses a study of phenotypes, but different, in that the phenotypes come from a heritable manipulation of the expression of DNA, not the DNA itself. This is a relatively new field that links heredity and developmental changes allowing for a deeper understanding of the ongoing changes occurring in organisms that cannot be accounted for by the DNA sequence. Figure 1 explains the difference between genetic and epigenetic changes affecting the phenotype.
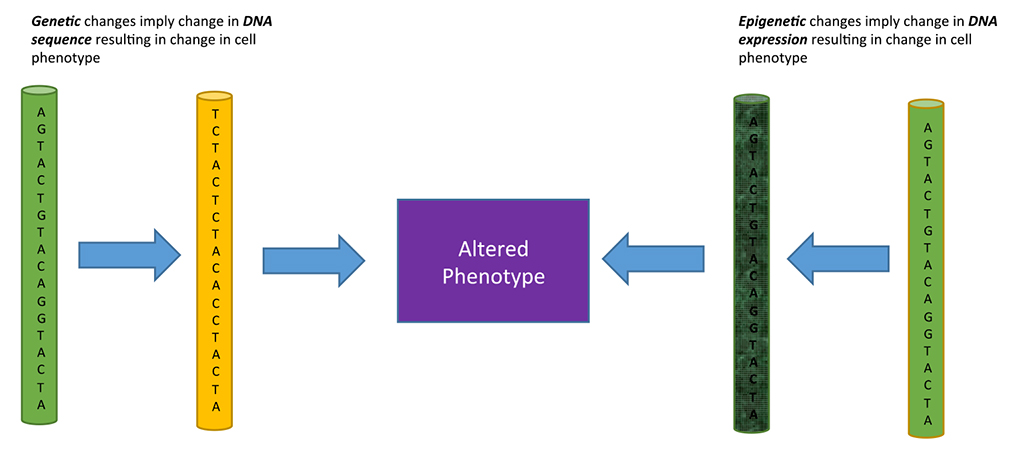
Figure 1. Genetic versus Epigenetic changes affecting phenotype
Methods and Materials
A descriptive review of the various aspects of epigenetics in the context of cancer was compiled using published literature sources. This is a vast subject and our review is a concise description of basic aspects of the role of epigenetic processes in cancer.
History of Epigenetics and its role in cancer
Epigenetics is a relatively new field of genetics, with the first glimpse into it occurring in 1878, when Walther Flemming began researching organisms and their phenotypes. Flemming discovered chromosomes and the process of mitosis during cell division. He focused on cytogenetics, the study of inheriting genes and traits based on the structure of chromosomes [1]. This, combined with Gregor Mendel’s work in genetics formed a foundation for understanding of genotypes and phenotypes. At this point, many researchers started to look at how, besides genetic makeup, there was another layer (epi) to the traits and characteristics an organism has. In the 1940’s, Conrad Waddington used the term “epigenetic landscape” to represent the layer of processes that occur within cells to change how DNA is expressed to decide the fate of cellular phenotype without affecting the genotype. This process is famously visualized as a ball rolling down one of various trajectories in an undulating landscape, called Waddinton’s Classical Epigenetic Landscape [2]. Interestingly enough, researchers began comparing epigenetics to the Lamarckian evolution. Lamarck’s theory looks at how acquired traits can be passed down to upcoming generations, while epigenetics examines how “acquired traits”, or traits that occur from changes in DNA expression take place, and how they can be passed down to subsequent generations, a process referred to as ‘soft inheritance’. This may be due to heritable changes in the chromatin, such as DNA methylation, occurring as a result of environmental influences [3]. In 1983, epigenetics in relation to cancer began to call more attention, as progressively lower levels of DNA methylation was discovered to correlate significantly with progression of normal tissue to cancerous tissue and metastatic disease [4]. A few years later, portions of the DNA with clusters of cytosine and guanine nucleotides called as the CpG cluster, was shown to, when highly methylated, indicate inactive promoters. Hypermethylation of certain tumor suppressor genes was discovered suggesting a very strong link between epigenetic mechanisms and cancer [5, 6]. Enormous strides in both research and clinical trials in the field of cancer epigenetics have led to the development of many epigenetic therapies to target cancer.
Epigenetic Mechanisms Linked to Cancer
Several different epigenetic mechanisms are linked to cancer. Some prominent ones are DNA methylation, histone modification and chromatin remodeling. All of these mechanisms are present in healthy organisms, and play a prominent role in growth and development as well as adaptation to the environment.
The influence of any one of these can result in many potential health problems ranging from increased risk of obesity to cancer. For example, during the Dutch Famine of 1944-1945, pregnant mothers who were not getting enough nutrition incurred less DNA methylation of the insulin-like growth factor II (IGF2) gene promoter in their early phase embryos. This prenatal exposure of embryos persisted into adulthood as a heritable epigenetic change [7]. It is important to understand that epigenetic factors can work in numerous ways, with cancer causing factors being a pivotal research topic. Environmental influences, such as eating certain kinds of foods and exposure to carcinogens can contribute to epigenetic factors causing cancer. Unlike genetic changes, epigenetic processes are targetable and can be reversed, thus offering avenues for prevention and control of cancers.
DNA Methylation and cancer
The most well-known epigenetic mechanism is DNA methylation. This occurs when a methyl group (CH3) from S-adenosylmethionine is added to a cytosine nucleotide by DNA methyltransferase (DNMT) enzymes [8]. When a section is methylated it is typically turned “off ”, or sent to a section of the major groove in the DNA that makes it hard for transcriptional factors to bind promoters and transcribe the gene. DNA hypermethylation is a process restricted to portions of DNA that are rich in CpG dinucleotides and are part of promoters of genes [9].
Consequently, many genes are hidden from transcription factors, and therefore not expressed. Repressing genes can result in cancer due to the number of tumor suppressor genes located in hypermethylated regions. Not only are tumor suppressor genes repressed, but also, other important genes that repair DNA and regulate the cell cycle are hypermethylated, causing cancers to both form and metastasize very quickly. The table below (Table 1) gives an overview of some of the pathways and genes affected by DNA hypermethylation and their relationship to various cancers.
Amongst the most prominent and earliest described epigenetic processes leading to cancer is low level of DNA methylation also called as DNA hypomethylation. The demethylation of repetitive DNA sequences, coding regions and introns, and gene poor areas are noted to be hallmarks of carcinogenesis. These processes result in chromosomal instability, reactivation of transposable elements and loss of imprinting. It leads to mitotic recombination leading to deletions and translocations as well as chromosomal rearrangements [10]. It can lead to reactivation of repetitive DNA sequences called retrotransposons an example of which is Long Interspersed Nuclear Element-1 (LINE1). LINE-1 are implicated in carcinogenesis of epithelial tumors [11]. These are genes that comprise around 17% of the human DNA are also called “jumping genes”. They propagate themselves throughout the genome via RNA transcription and reverse transcription back into the genome and are thus called retrotransposons. DNA methylation keeps these jumping genes in check in normal cells. Hypomethylation leads to activation of these transposable genes. By inserting themselves in proximity to key genes they lead to activation of oncogenic signaling pathways [12].
Table 1. Pathways affected by DNA hypermethylation in cancer
| Pathway Affected | Gene | Cancer | Reference |
| DNA repair pathway | BRCA1 | Breast , ovarian | [15] |
| MLH1 | GI (colon and gastric), endometrial, ovarian | [16] | |
| MGMT | Lung, brain | [17, 18] | |
| Cell cycle checkpoint | CDKN2A Colorectal | [19] | |
| p15 | Bladder | [20] | |
| RASSF1A | Breast, lung | [21] | |
| Cytoproliferative and metastasis pathways | Ecadherin Ovarian, |
breast | [22] |
| APC | Colorectal | [23] | |
Research suggests that hypomethylation results in activation of oncogenes, allowing cancers to form and spread rapidly. An example of this is prostate cancer, where the promoter for gene LINE1 is found to be significantly hypomethylated. This epigenetic process marks the progress of prostate cancer from localized disease to metastatic disease [13]. Global hypomethylation can affect certain tissues in the body that then go on to affect how LINE1 is transcribed. In fact, hypomethylation has been shown to result in a specific biologic signature, which marks invasiveness, and is common to breast cancer, prostate cancer and liver cancer. It utilizes major pathways such as TGF-β (transforming growth factor-beta) and ERBB2 triggered pathways [14-23].
Histone Modifications and cancer
While DNA methylation is a huge component of epigenetics; there are many other mechanisms at play. One such mechanism is chromatin remodeling which is responsible for many cancers, diseases, and conditions. To understand chromatin remodeling and cancer, it is first important to look at histones and how they affect gene expression. Histones are alkaline proteins that DNA wraps around to form nucleosomes. Nucleosomes are then tightly coiled to form chromatin, and subsequently chromatids. Naturally, because they are in such close contact with the DNA, histones play a key role in gene expression regulation. The N-terminal tail of histones undergoes posttranslational modifications (PTMs), thereby affecting the accessibility of DNA to RNA polymerase. The N-terminal tail can be changed by methylation, acetylation, ubiquitylation, and phosphorylation [24, 25]. Histone acyteltransferases (HAT’s), histone methyl transferases (HMT’s), Histone deacetylases (HDACs) and histone demethylases (KDMs) are some histone modifying enzymes performing these PTMs. An acetylated histone is comparatively looser than a normal histone, allowing for many transcription sites to become available to RNA polymerases. Increases in gene activation follow this process. Histone deacytelases (HDAC’s) support the removal of acetyl groups from the lysine amino acid in histones, allowing the DNA to tightly wrap around the histone. This results in gene silencing, as many promoter regions are hidden and cannot be transcribed. Obviously, any significant changes or mistakes in PTM’s can easily cause repression of important genes and activation of situationally harmful genes. For example, histone deacetylases act to repress the transcription of tumor suppressor genes.
Additionally, histone variants and histone chaperones are other players in epigenetic processes affecting gene expression. Histone modification processes are important for normal cellular development but can be hijacked by cancer for oncogenic signaling. This may be due to an altered histone code leading to aberrant gene expression or mutations in enzymes regulating histone modifications[26-30].
Examples of histone-mediated processes involved in cancer are presented in Table 2.
Table 2. Histone modifications in cancer
| Histone modification | Cancer | Reference |
| Histone variant H3.3 mutations | Glioblastoma | [27] |
| SETD2 and KDM5C inactivating mutations | Renal Cell Carcinoma | [28] |
| Downregulation of H4K16ac and H4K20me3 |
Colorectal cancer and leukemia | [29] |
| H3K27me3 inactivates tumor suppressor genes | Prostate Cancer and breast cancer | [30] |
Chromatin Remodeling and Cancer
Chromatin remodeling is a dynamic process that facilitates the remodeling of nucleosomes. Chromatin modification plays an essential role in DNA replication and transcription. Tightly packaged chromatin is called heterochromatin, while loosely packaged chromatin is called euchromatin. There are four well described chromatin remodeling complexes: SWI/SNF, ISWI, INO80 and NuRD/Mi-2. By means of some key characteristic features, these remodeling complexes are able to interact with the nucleosome core, use energy from ATP, bind histones and provide grounds for biochemical alteration and protein-protein interactions [31]. For example, the switch/sucrose non-fermentable chromatin remodeling complexes (SWI/SNF) are responsible for transcription control – as well as a multitude of other tasks such as DNA repair and chromosome segregation [32]. Mutations in subunits of the SWI/SNF complex are known to occur in cancer. Malignant rhabdoid tumors are the result of the loss of INI1 subunit of the SWI/SNF complex [33].
Noncoding RNAs and Cancer
Noncoding RNAs are RNAs that are not translated into protein but play a role in gene expression and translation by various mechanims both in the normal cell and cancer. Long non-coding RNAs (lncRNAs) are non-coding regulatory sequences that have a role to play in gene expression. These are implicated in tumorigenesis by various mechanisms. They may remodel chromatin, act as transcriptional co-activators or repressors, inhibit protein, post transcriptional modifications or serve as decoy elements. Single nucleotide polymorphisms (SNPs) in lncRNAs are implicated in the heritability of several cancers including breast, thyroid, prostate and ovarian [34].
Micro-RNAs (miRNA) are short non-coding RNAs that regulate translation of transcribed genes and hence influence the phenotype of a cell. miRNA mediated pathways are implicated in several cancers including glioblastoma [35].
Table 3 summarizes the various epigenetic processes presented in this review.
Table 3. Epigenetic processes implicated in cancer
| Process: | Function: |
| DNA Methylation | Used to control gene expression by methylated specific parts of DNA, therefore turning those parts “on” or “off ” |
| Histone Modifications | A post transitional modifications made to histones that change which aspects of the DNA are available to be promoter regions |
| Chromatin Remodeling | Packaging or loosening up chromatin, allowing DNA to be easily read, or tightening the chromatin so that DNA is more difficult to be read. Both result in changes to promoter regions. |
| Non – coding RNA | A RNA that is not translated into a protein, and instead regulations gene expression and translation |
General Epigenetic Therapies
Several epigenetic therapies have been developed and approved for use in various malignancies. Several novel epigenetic therapies are in the process of preclinical and clinical development. DNMT inhibitors are the first epigenetic therapy approved for clinical use for their therapeutic role in myelodysplatic syndrome and acute myeloid leukemia,
HDAC inhibitors are approved for use in T cell lymphoma and multiple myeloma.
Several other epigenetic targets have been identified and their inhibitors are in preclinical and clinical development. These include bromodomain, EZH2 (enhancer of zeste homolog 2) and DOT1( disrupter of telomere silencing protein 1) inhibitors [36].
Conclusion
The role of epigenetics in cancer is prominent and clear. This relationship is apparent through the multiple epigenetic mechanisms implicated in cancer: DNA methylation, histone modifications, chromatin remodeling, and non-coding RNA. All of these mechanisms have been proven to significantly impact the epigenome, without impacting the DNA itself. DNA methylation can occur in a surplus or a deficit, both of which can consequently create and promote cancer within the body. Histone modifications alter the space in which DNA resides, therefore making it harder or easier for that
DNA to be transcribed. Chromatin remodeling affects the degree to which specific sites of DNA are bound, which decides to what extent certain genes are expressed. Long non-coding RNA is a more recently discovered major epigenetic mechanism regulating gene expression. While epigenetic mechanisms can result in cancer formation, research underway holds promise of using the same mechanisms to combat cancer.
Acknowledgements: The authors would like to thank Dr. Sunil Sharma and the Center for Investigational Therapeutics at Huntsman Cancer Institute for their support and providing the opportunity to learn.
Funding information: Support for the publication of this manuscript was provided by the Huntsman Cancer Institute/Huntsman Cancer Foundation.
Conflict of Interest: The authors have no competing interests with regard to this review.
References
- Paweletz N (2001) Walther Flemming: pioneer of mitosis research. Nat Rev MolCell Biol 2: 72–75. [crossref]
- Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128: 635–638. [crossref]
- Jablonka E and MJ Lamb (2008) Soft inheritance: challenging the modern synthesis. Genetics and Molecular Biology 31: 389–395.
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, et al. (1983) The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 11: 6883-6894. [crossref]
- Baylin SB, Höppener JW, de Bustros A, Steenbergh PH, Lips CJ, et al. (1986) DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res 46: 2917–2922. [crossref]
- Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4: 143–153. [crossref]
- Heijmans BT, et al. (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences 105: 17046–17049.
- Das PM, Singal R (2004) DNA methylation and cancer. J Clin Oncol 22: 4632–4642. [crossref]
- Kulis M, Esteller M (2010) DNA methylation and cancer. Adv Genet 70: 27–56. [crossref]
- Feinberg AP and Vogelstein B (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92.
- Carreira PE, Richardson SR, Faulkner GJ (2014) L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J 281: 63–73. [crossref]
- Xiao-Jie L, et al. (2016) LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med 18: 431–439.
- Yegnasubramanian S, et al. (2008) DNA Hypomethylation Arises Later in Prostate Cancer Progression than CpG Island Hypermethylation and Contributes to Metastatic Tumor Heterogeneity. Cancer Research 68: 8954–8967.
- Cheishvili D, et al. (2015) A common promoter hypomethylation signature in invasive breast, liver and prostate cancer cell lines reveals novel targets involved in cancer invasiveness. Oncotarget 6: 33253–33268.
- Hansmann T, et al. (2012) Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Human Molecular Genetics 21: 4669–4679.
- Richman S (2015) Deficient mismatch repair: Read all about it (Review). Int J Oncol 47: 1189–1202. [crossref]
- Gömöri É, et al. (2012) Concurrent hypermethylation of DNMT, MGMT and EGFR genes in progression of gliomas. Diagnostic Pathology 7: 8–8.
- Gu C, Lu J, Cui T, Lu C, Shi H, et al. (2013) Association between MGMT promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One 8: e72633. [crossref]
- Shima K, et al. (2011) Prognostic Significance of CDKN2A (p16) Promoter Methylation and Loss of Expression in 902 Colorectal Cancers: Cohort Study and
Literature Review. International journal of cancer. Journal international du cancer 128: 1080–1094. - Bilgrami SM, et al. (2014) Promoter hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. SpringerPlus 3: 178.
- Grawenda AM, E O’Neill (2015) Clinical utility of RASSF1A methylation in human malignancies. British Journal of Cancer 113: 372–381.
- Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ (2012) E-cadherin’s dark side: possible role in tumor progression. Biochimica et Biophysica Acta 1826: 23–31.
- Liang TJ, Wang HX2, Zheng YY3, Cao YQ4, Wu X5, et al. (2017) APC hypermethylation for early diagnosis of colorectal cancer: a meta-analysis and literature review. Oncotarget 8: 46468–46479. [crossref]
- Morales V, Richard-Foy H (2000) Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol Cell Biol 20: 7230–7237. [crossref]
- Venkatesh S, Workman JL (2015) Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16: 178–189. [crossref]
- Hattori N and Ushijima T (2014) Compendium of aberrant DNA methylation and histone modifications in cancer. Biochemical and Biophysical Research Communications 455: 3–9.
- Schwartzentruber J, et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231.
- Dalgliesh GL, et al. (2010) Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363.
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400. [crossref]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, et al. (2008) Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750. [crossref]
- Nair SS, Kumar R (2012) Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol Oncol 6: 611–619. [crossref]
- Euskirchen GM, et al. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008.
- Kordes U, Gesk S, Frühwald MC, Graf N, Leuschner I, et al. (2010) Clinical and molecular features in patients with atypical teratoid rhabdoid tumor or malignant rhabdoid tumor. Genes Chromosomes Cancer 49: 176–181. [crossref]
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME (2013) Long noncoding RNAs and the genetics of cancer. Br J Cancer 108: 2419–2425. [crossref]
- Katsushima K, Kondo Y (2014) Non-coding RNAs as epigenetic regulator of glioma stem-like cell differentiation. Front Genet 5: 14. [crossref]
- Jones PA, Issa JP, Baylin S (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17: 630–641. [crossref]